Summary information and primary citation
- PDB-id
- 2ewj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication-DNA
- Method
- X-ray (2.7 Å)
- Summary
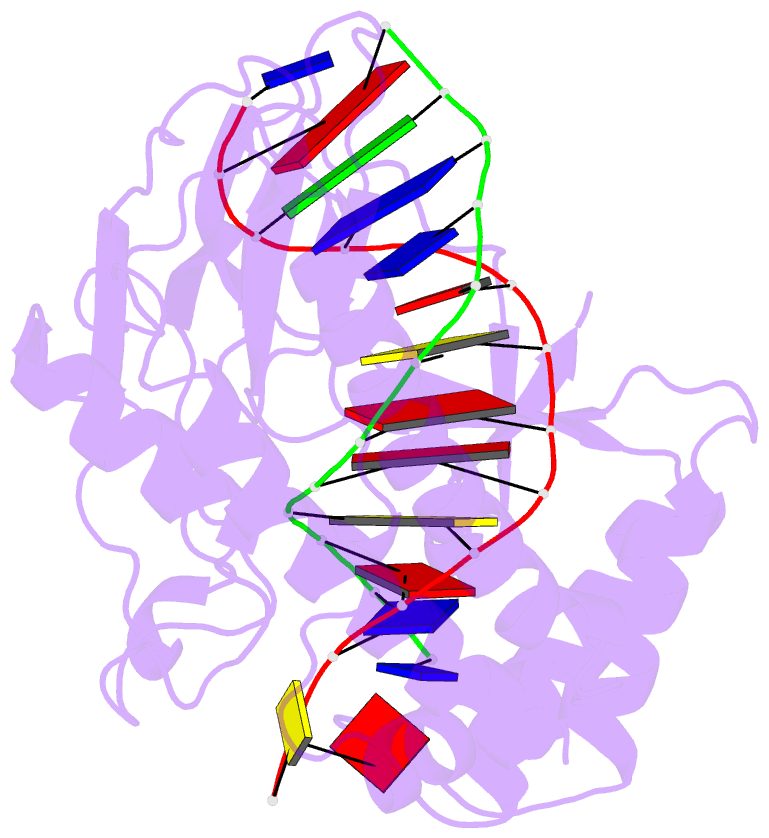
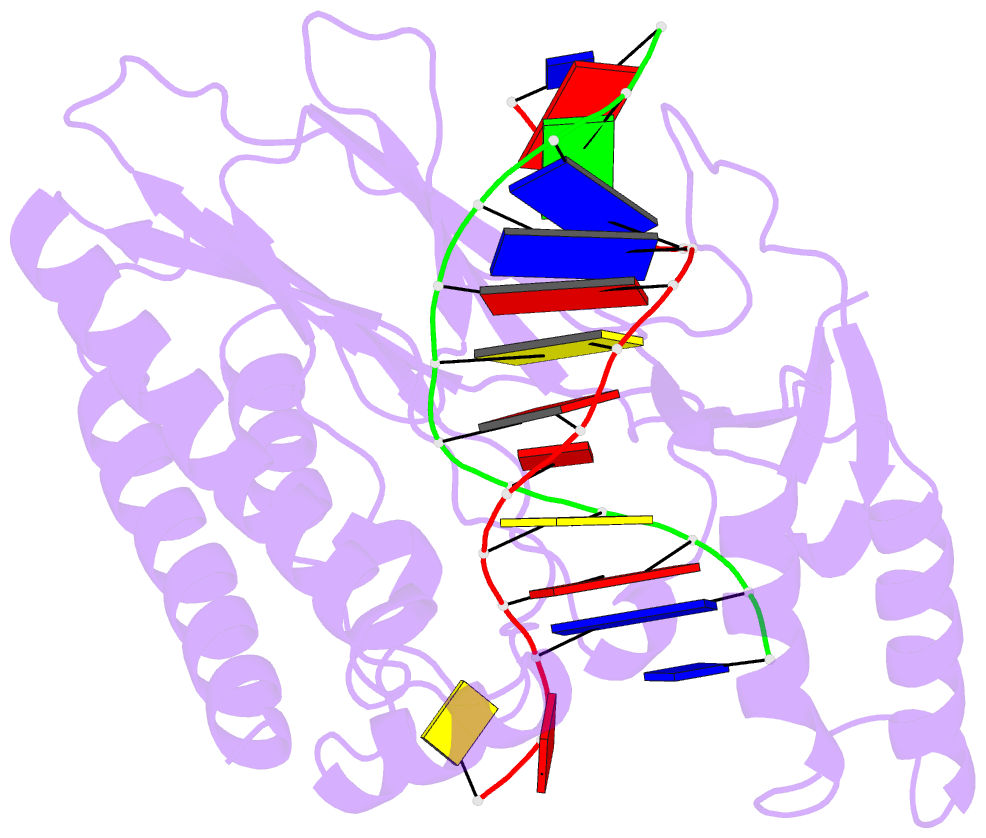
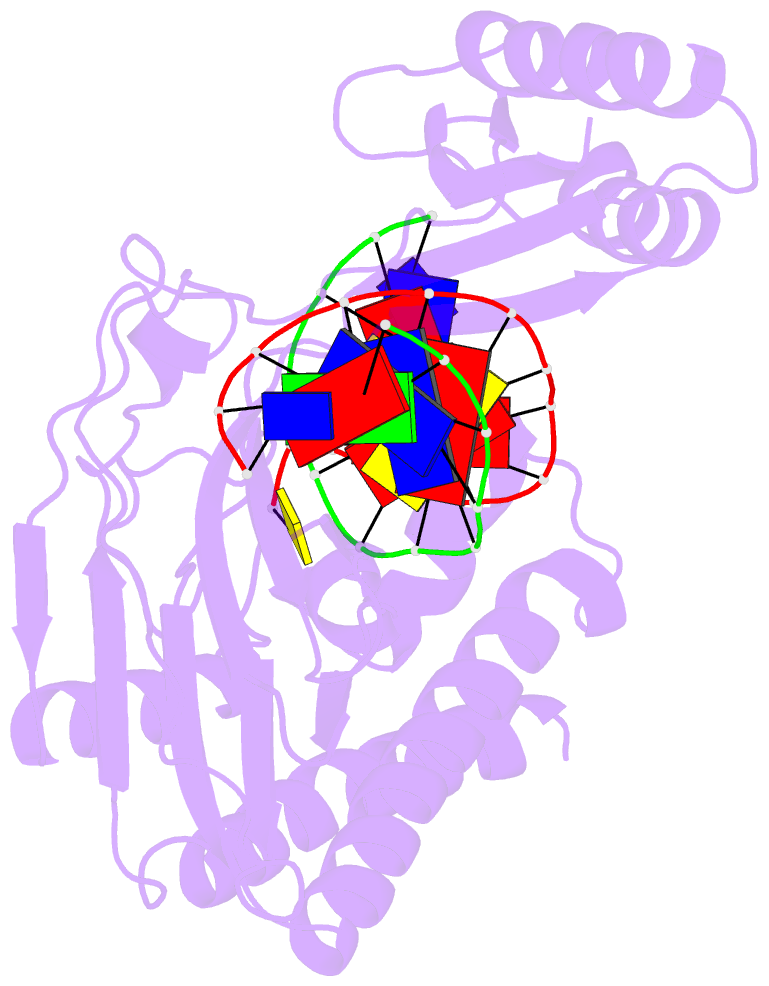
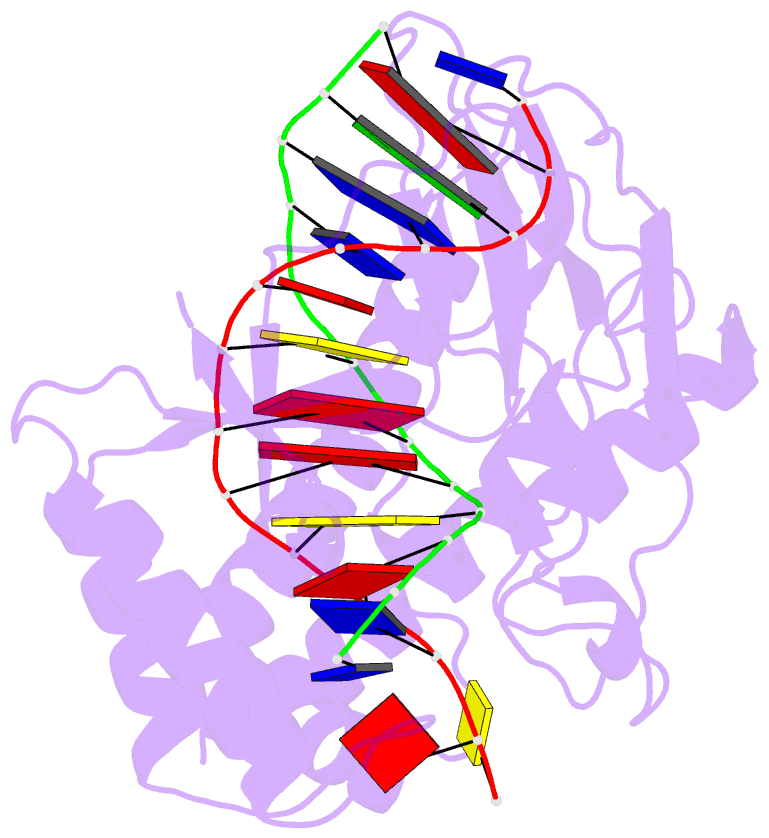
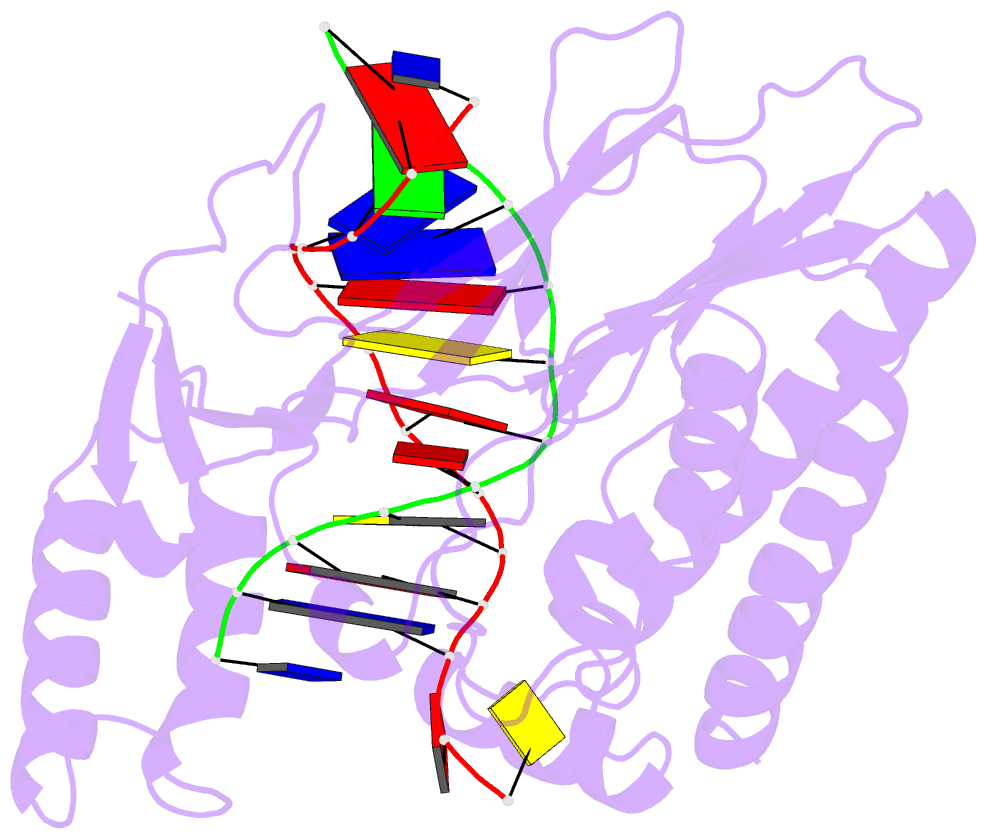
- Escherichia coli replication terminator protein (tus) complexed with DNA- locked form
- Reference
- Mulcair MD, Schaeffer PM, Oakley AJ, Cross HF, Neylon C, Hill TM, Dixon NE (2006): "A molecular mousetrap determines polarity of termination of DNA replication in E. coli." Cell(Cambridge,Mass.), 125, 1309-1319. doi: 10.1016/j.cell.2006.04.040.
- Abstract
- During chromosome synthesis in Escherichia coli, replication forks are blocked by Tus bound Ter sites on approach from one direction but not the other. To study the basis of this polarity, we measured the rates of dissociation of Tus from forked TerB oligonucleotides, such as would be produced by the replicative DnaB helicase at both the fork-blocking (nonpermissive) and permissive ends of the Ter site. Strand separation of a few nucleotides at the permissive end was sufficient to force rapid dissociation of Tus to allow fork progression. In contrast, strand separation extending to and including the strictly conserved G-C(6) base pair at the nonpermissive end led to formation of a stable locked complex. Lock formation specifically requires the cytosine residue, C(6). The crystal structure of the locked complex showed that C(6) moves 14 A from its normal position to bind in a cytosine-specific pocket on the surface of Tus.