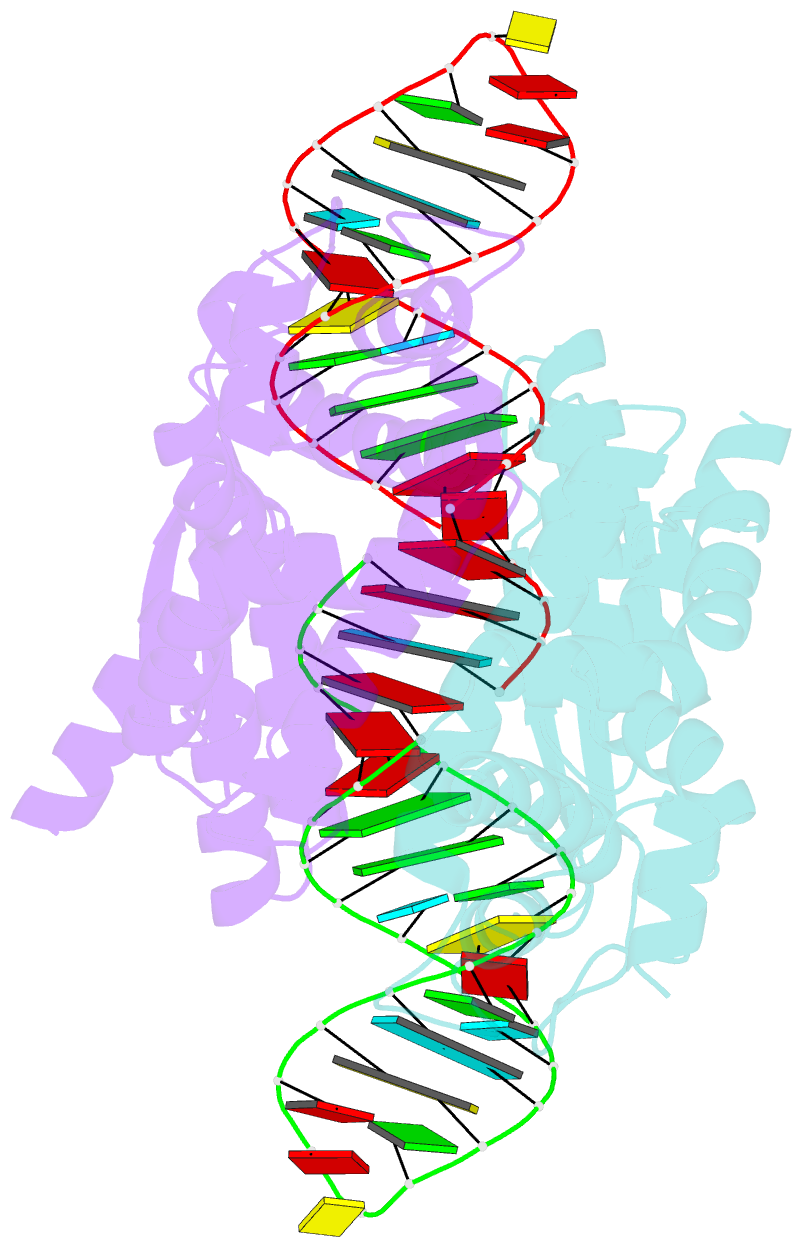
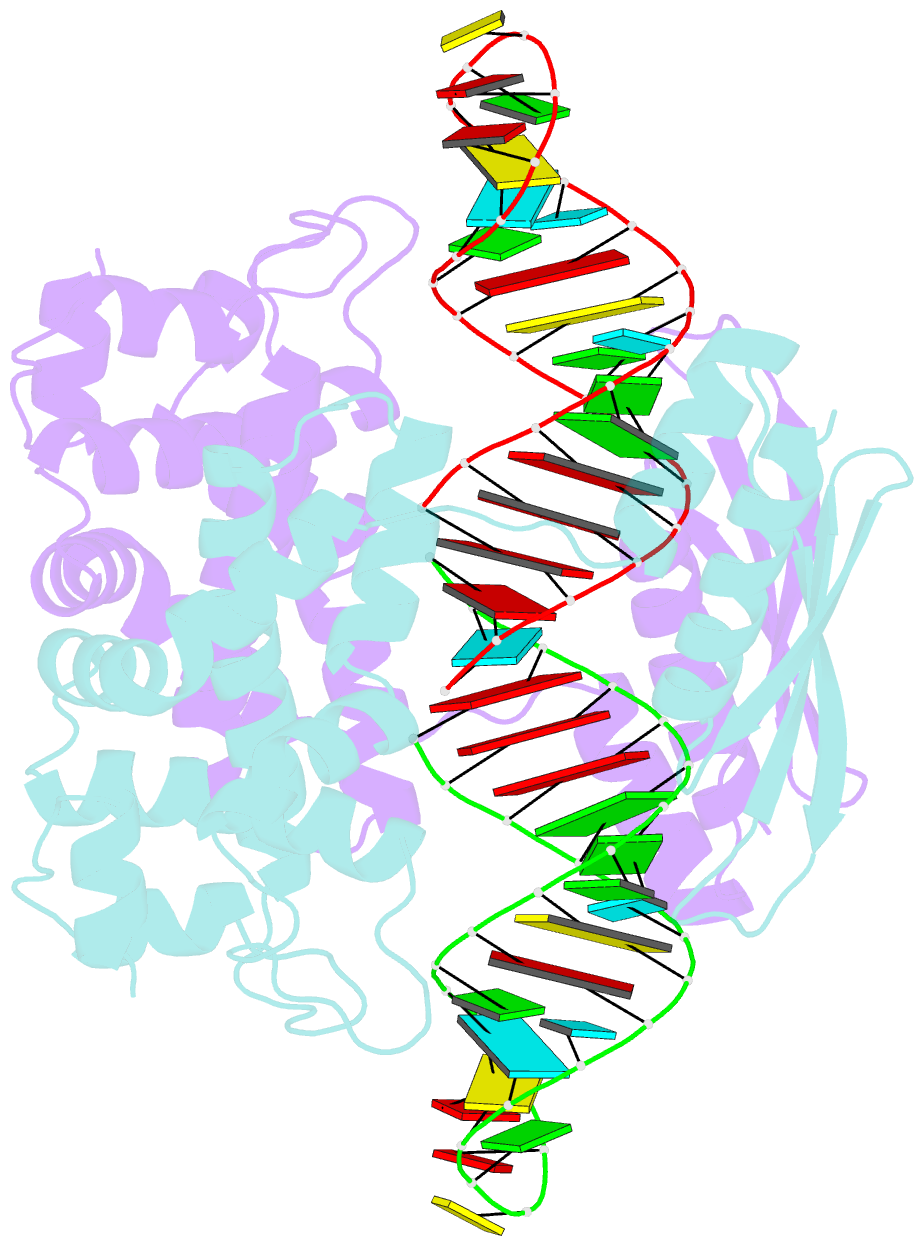
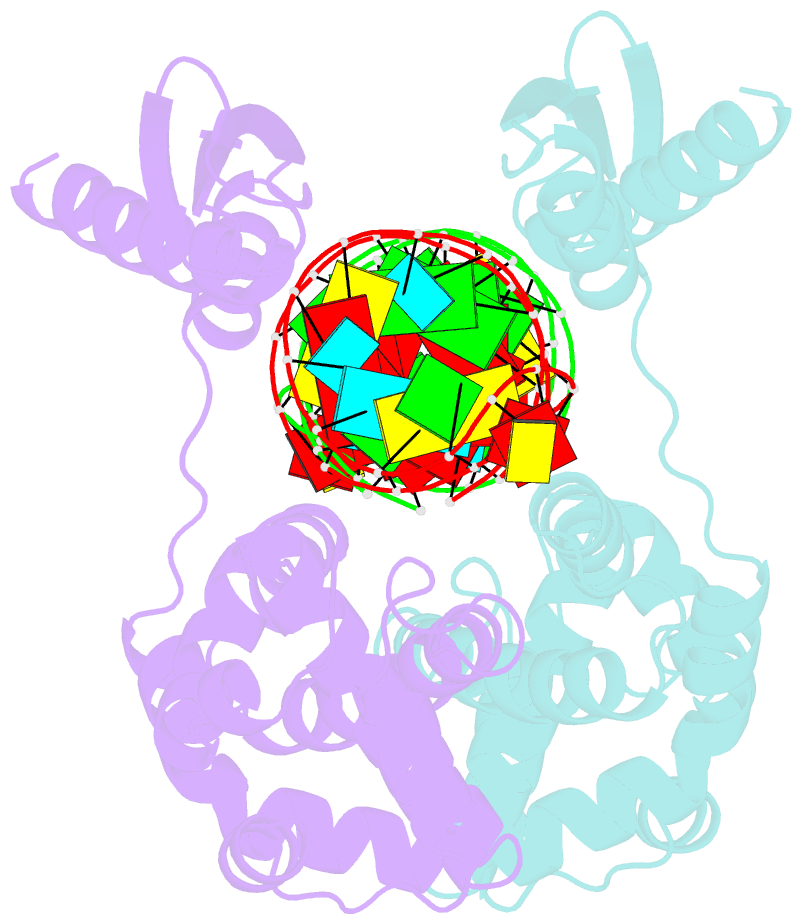
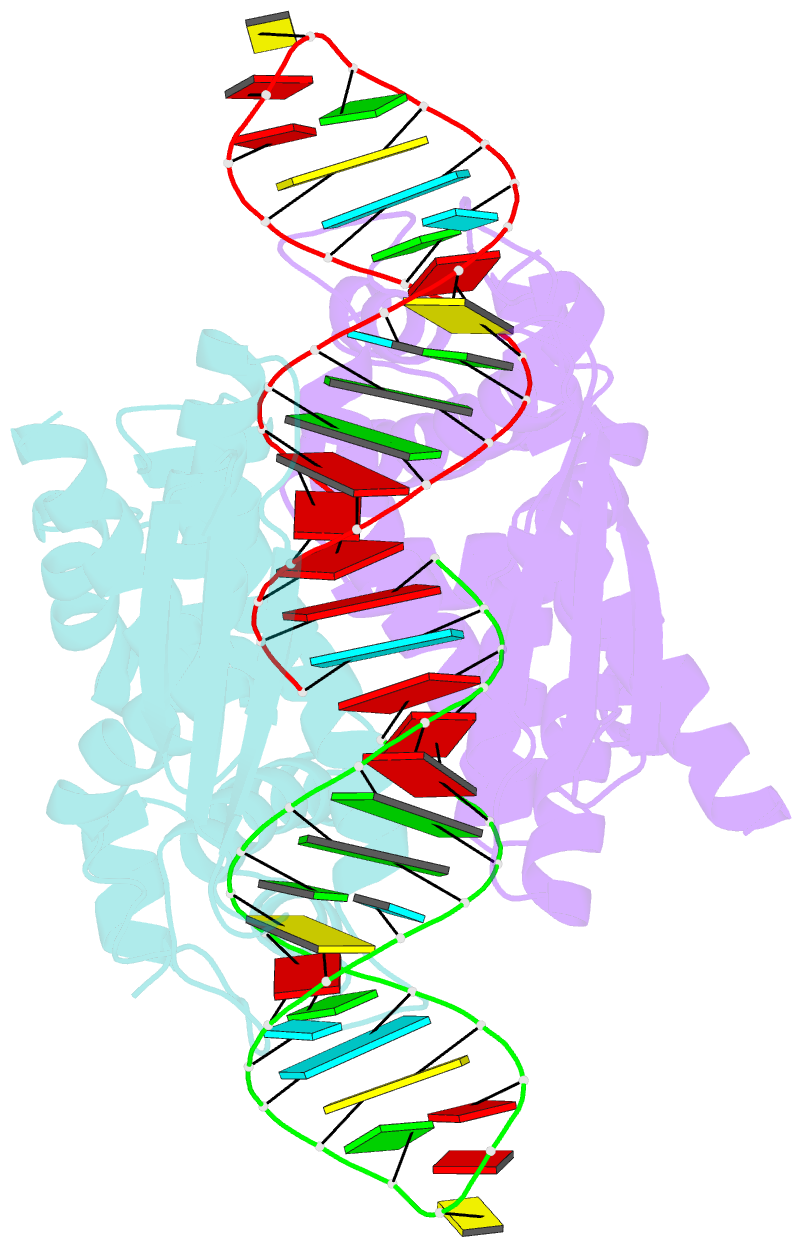
Summary information and primary citation
- PDB-id
- 2ez6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.05 Å)
- Summary
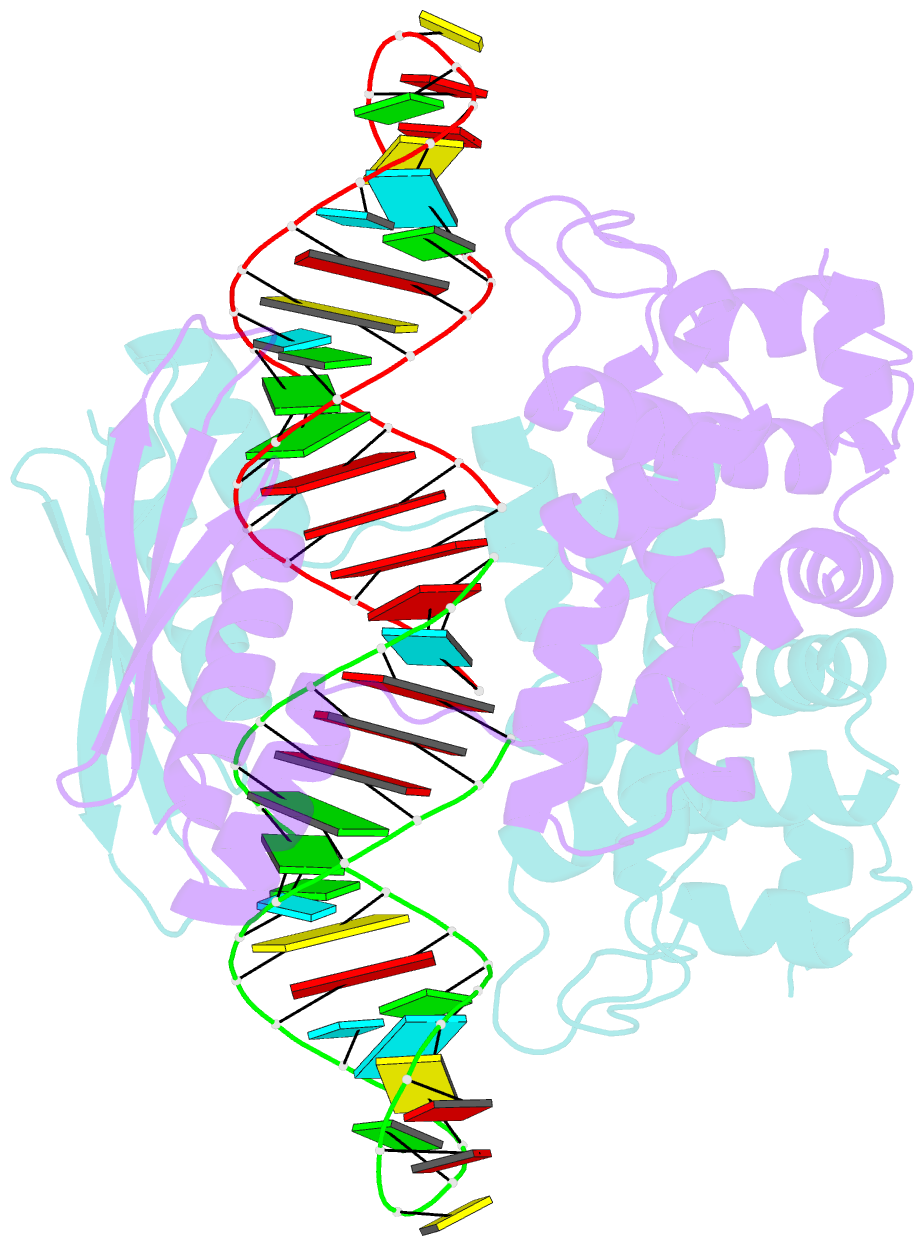
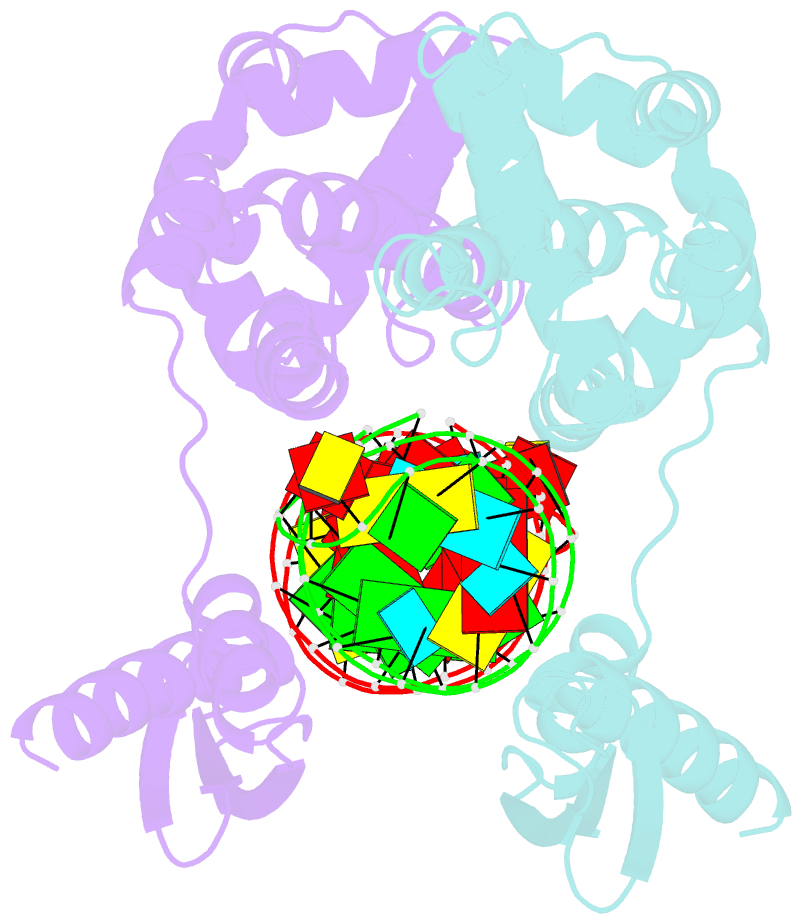
- Crystal structure of aquifex aeolicus rnase iii (d44n) complexed with product of double-stranded RNA processing
- Reference
- Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X (2006): "Structural Insight into the Mechanism of Double-Stranded RNA Processing by Ribonuclease III." Cell(Cambridge,Mass.), 124, 355-366. doi: 10.1016/j.cell.2005.11.034.
- Abstract
- Members of the ribonuclease III (RNase III) family are double-stranded RNA (dsRNA) specific endoribonucleases characterized by a signature motif in their active centers and a two-base 3' overhang in their products. While Dicer, which produces small interfering RNAs, is currently the focus of intense interest, the structurally simpler bacterial RNase III serves as a paradigm for the entire family. Here, we present the crystal structure of an RNase III-product complex, the first catalytic complex observed for the family. A 7 residue linker within the protein facilitates induced fit in protein-RNA recognition. A pattern of protein-RNA interactions, defined by four RNA binding motifs in RNase III and three protein-interacting boxes in dsRNA, is responsible for substrate specificity, while conserved amino acid residues and divalent cations are responsible for scissile-bond cleavage. The structure reveals a wealth of information about the mechanism of RNA hydrolysis that can be extrapolated to other RNase III family members.