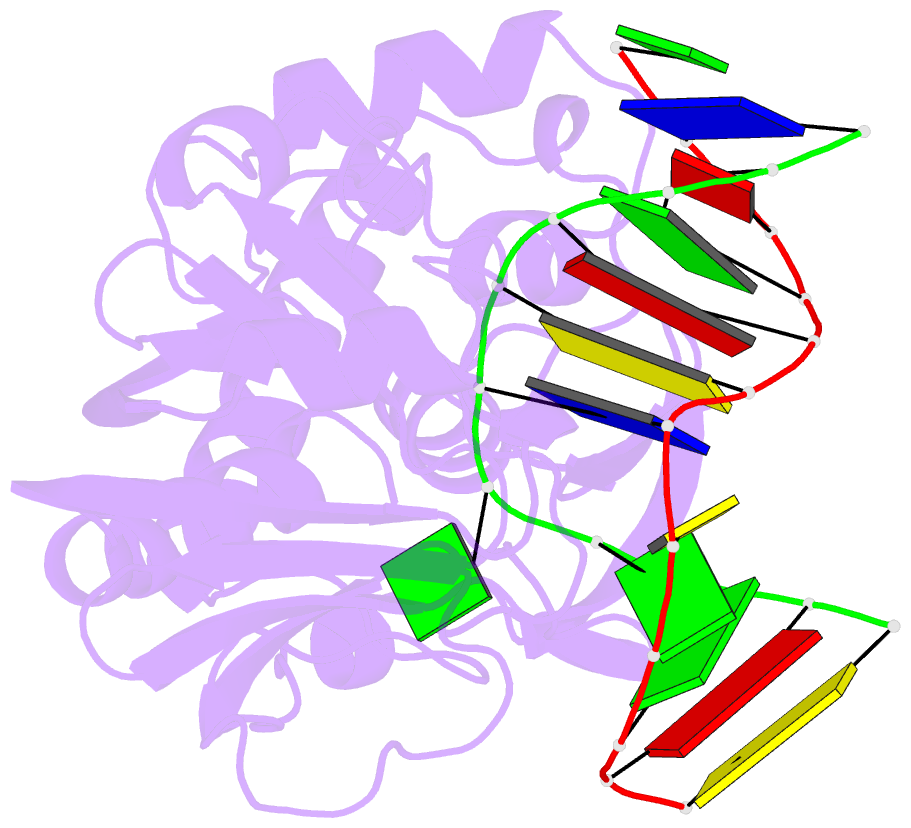
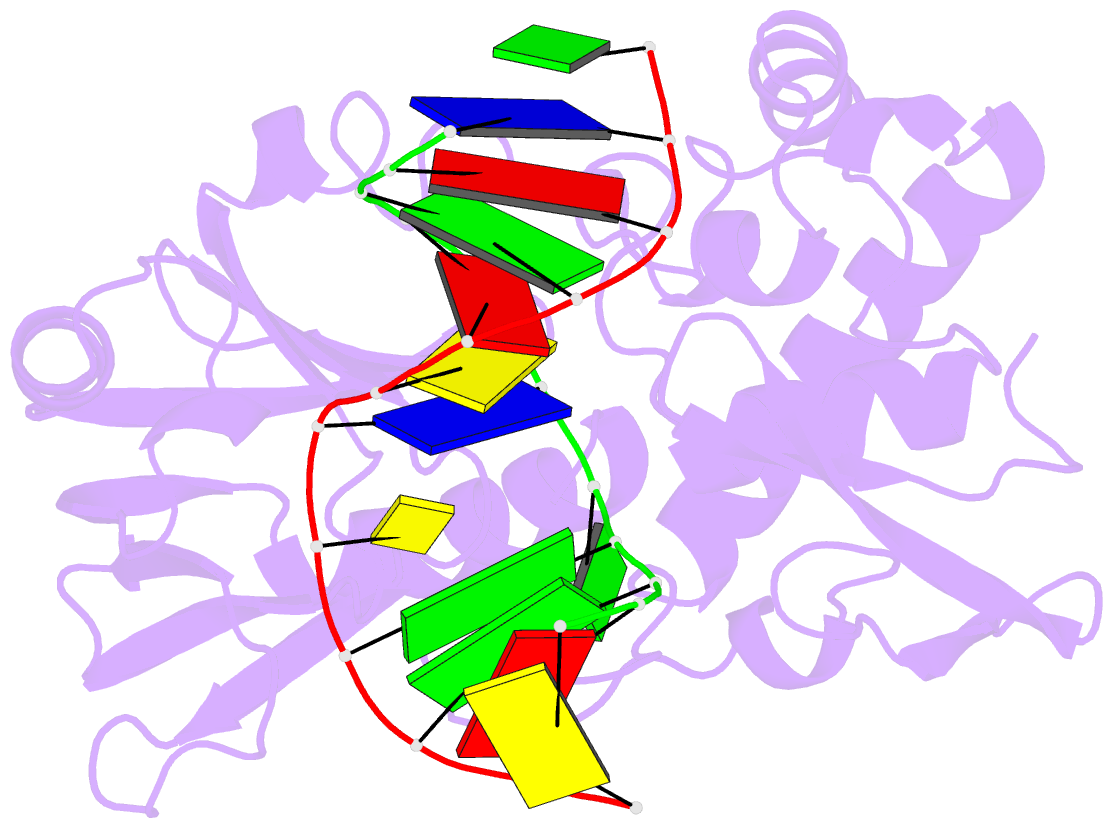
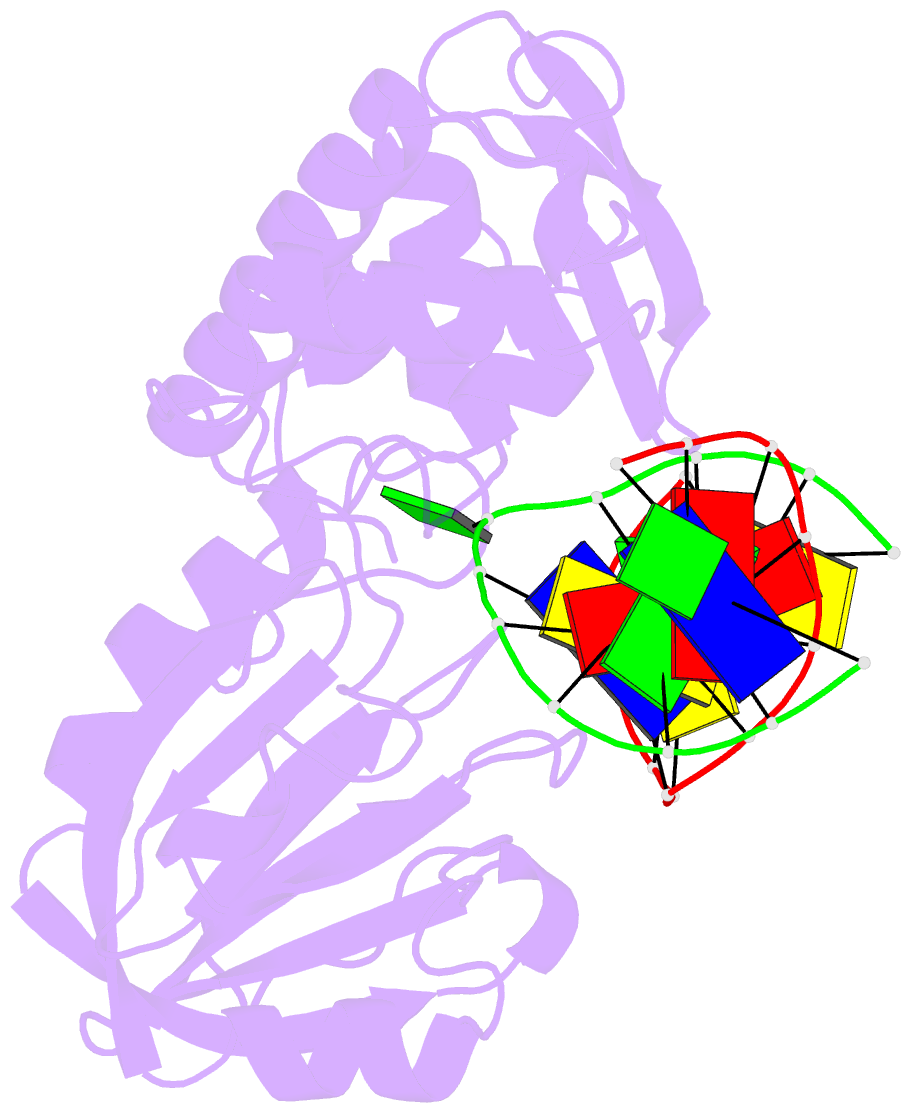
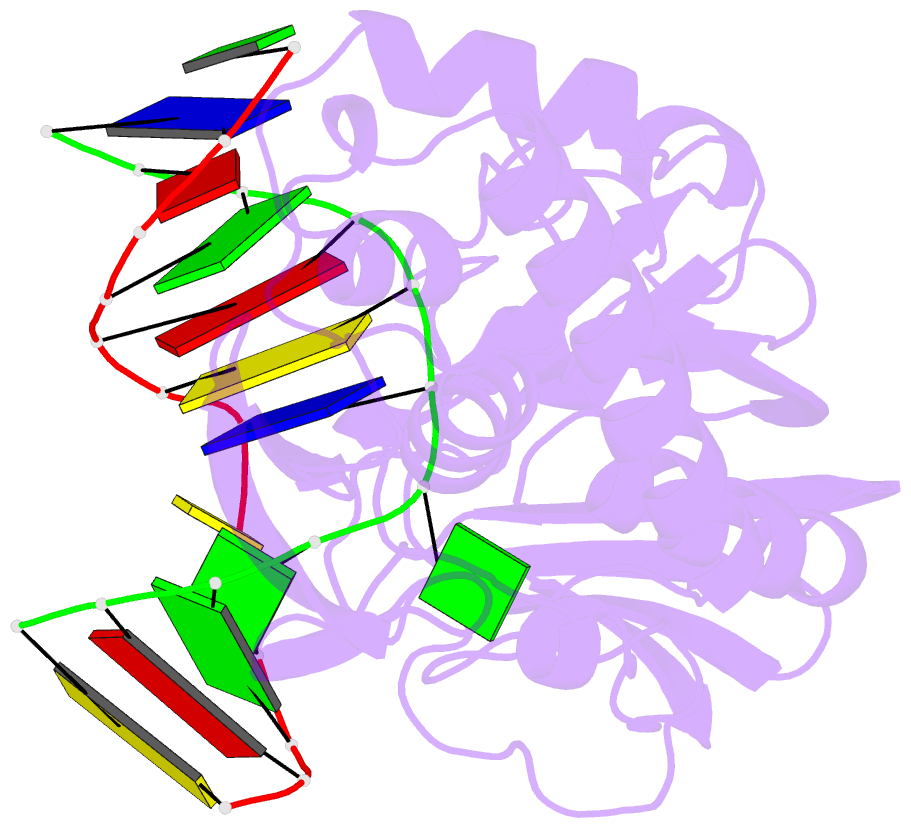
Summary information and primary citation
- PDB-id
- 2f5s; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.35 Å)
- Summary
- Catalytically inactive (e3q) mutm crosslinked to oxog:c containing DNA cc1
- Reference
- Banerjee A, Santos WL, Verdine GL (2006): "Structure of a DNA glycosylase searching for lesions." Science, 311, 1153-1157. doi: 10.1126/science.1120288.
- Abstract
- DNA glycosylases must interrogate millions of base pairs of undamaged DNA in order to locate and then excise one damaged nucleobase. The nature of this search process remains poorly understood. Here we report the use of disulfide cross-linking (DXL) technology to obtain structures of a bacterial DNA glycosylase, MutM, interrogating undamaged DNA. These structures, solved to 2.0 angstrom resolution, reveal the nature of the search process: The protein inserts a probe residue into the helical stack and severely buckles the target base pair, which remains intrahelical. MutM therefore actively interrogates the intact DNA helix while searching for damage.