Summary information and primary citation
- PDB-id
- 2fcc; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (2.3 Å)
- Summary
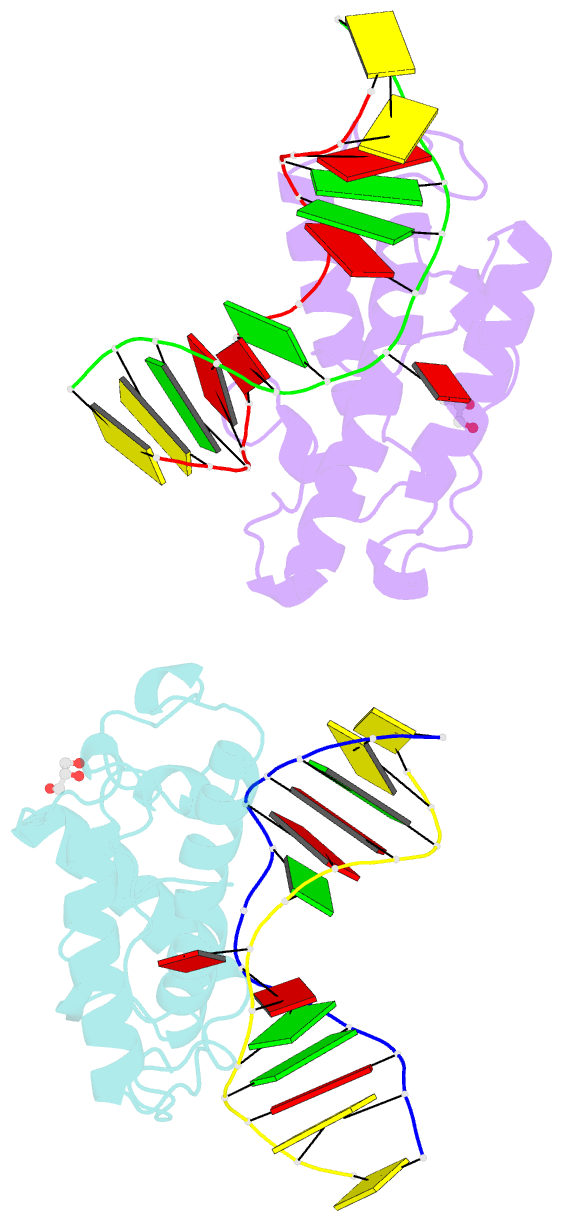
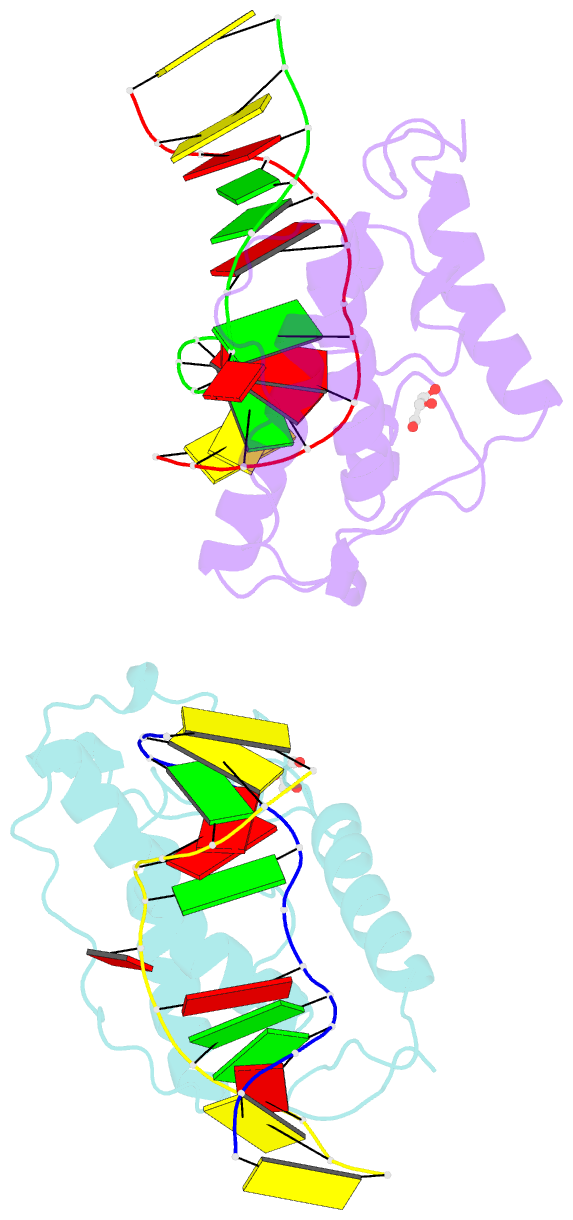
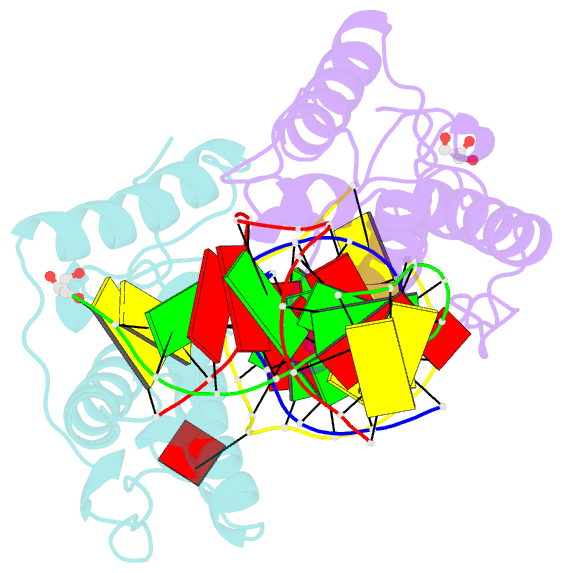
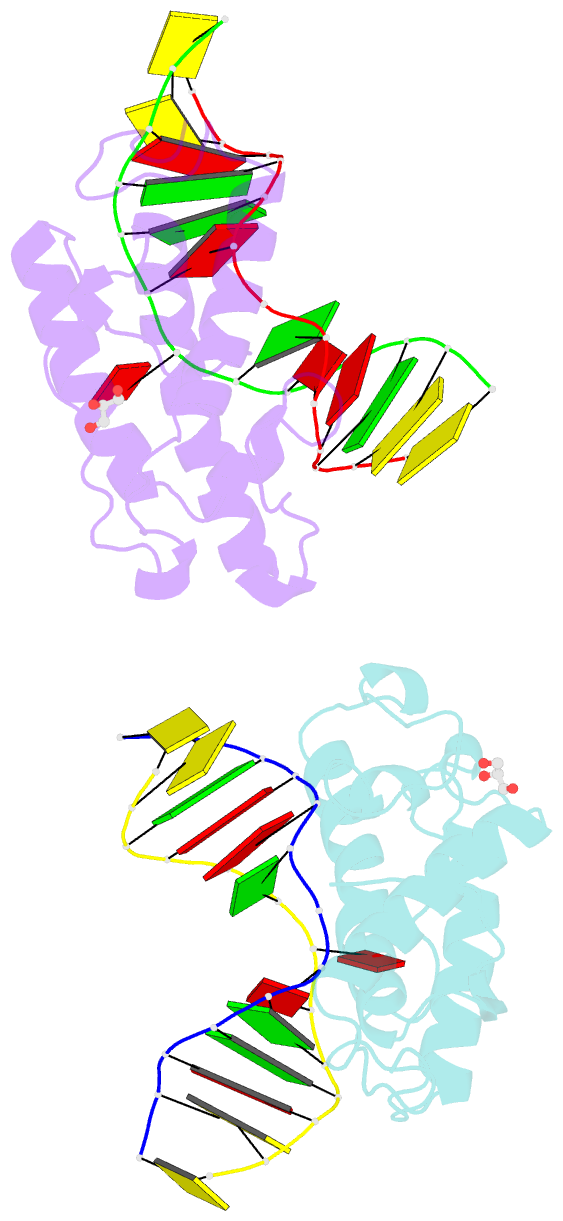
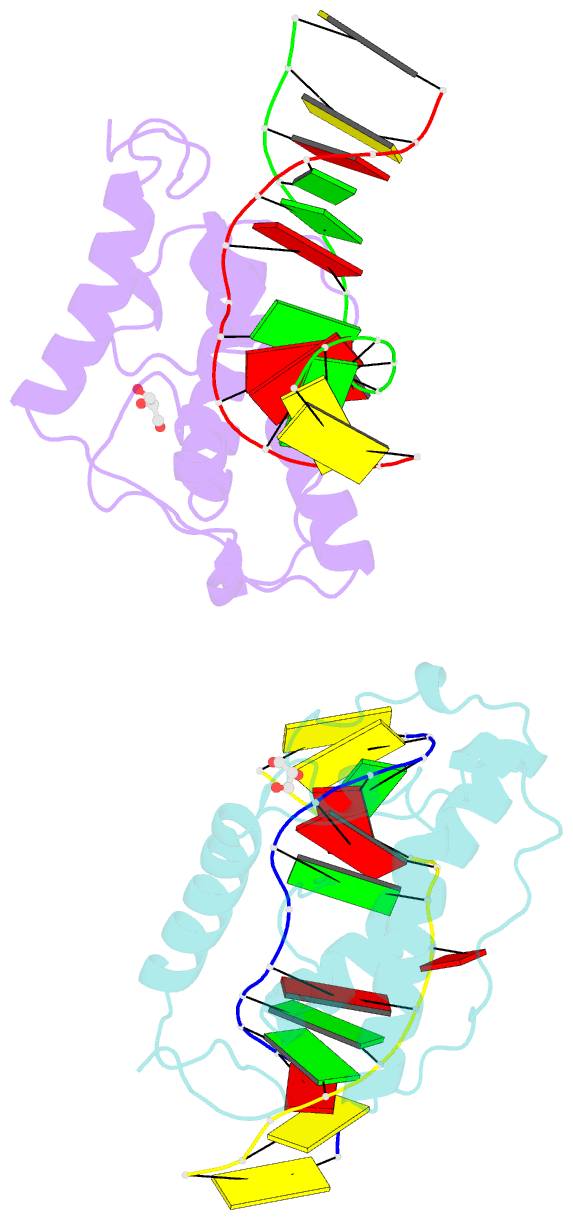
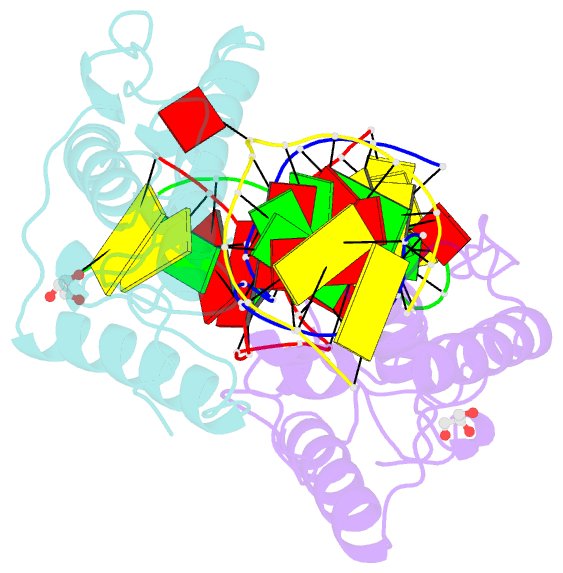
- Crystal structure of t4 pyrimidine dimer glycosylase (t4-pdg) covalently complexed with a DNA substrate containing abasic site
- Reference
- Golan G, Zharkov DO, Grollman AP, Dodson ML, McCullough AK, Lloyd RS, Shoham G (2006): "Structure of T4 Pyrimidine Dimer Glycosylase in a Reduced Imine Covalent Complex with Abasic Site-containing DNA." J.Mol.Biol., 362, 241-258. doi: 10.1016/j.jmb.2006.06.059.
- Abstract
- The base excision repair (BER) pathway for ultraviolet light (UV)-induced cyclobutane pyrimidine dimers is initiated by DNA glycosylases that also possess abasic (AP) site lyase activity. The prototypical enzyme known to catalyze these reactions is the T4 pyrimidine dimer glycosylase (T4-Pdg). The fundamental chemical reactions and the critical amino acids that lead to both glycosyl and phosphodiester bond scission are known. Catalysis proceeds via a protonated imine covalent intermediate between the alpha-amino group of the N-terminal threonine residue and the C1' of the deoxyribose sugar of the 5' pyrimidine at the dimer site. This covalent complex can be trapped as an irreversible, reduced cross-linked DNA-protein complex by incubation with a strong reducing agent. This active site trapping reaction is equally efficient on DNA substrates containing pyrimidine dimers or AP sites. Herein, we report the co-crystal structure of T4-Pdg as a reduced covalent complex with an AP site-containing duplex oligodeoxynucleotide. This high-resolution structure reveals essential precatalytic and catalytic features, including flipping of the nucleotide opposite the AP site, a sharp kink (approximately 66 degrees ) in the DNA at the dimer site and the covalent bond linking the enzyme to the DNA. Superposition of this structure with a previously published co-crystal structure of a catalytically incompetent mutant of T4-Pdg with cyclobutane dimer-containing DNA reveals new insights into the structural requirements and the mechanisms involved in DNA bending, nucleotide flipping and catalytic reaction.