Summary information and primary citation
- PDB-id
- 2fk6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.9 Å)
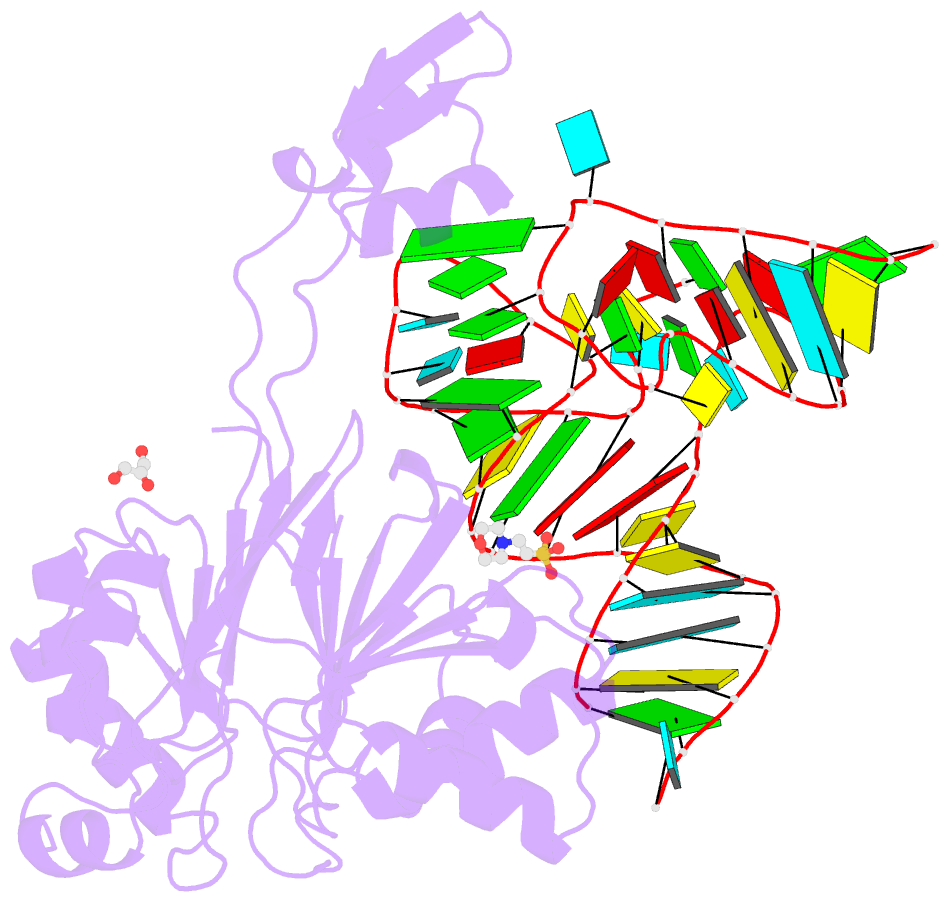
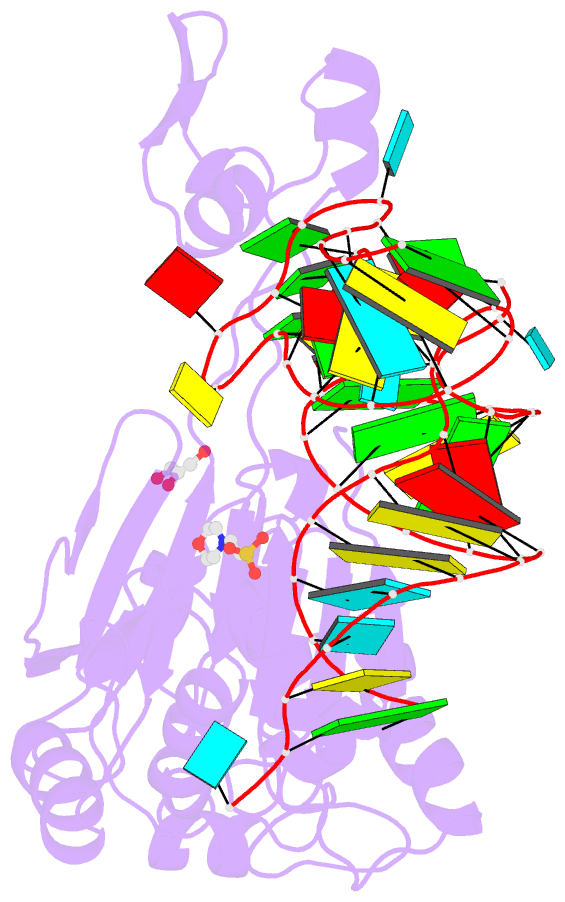
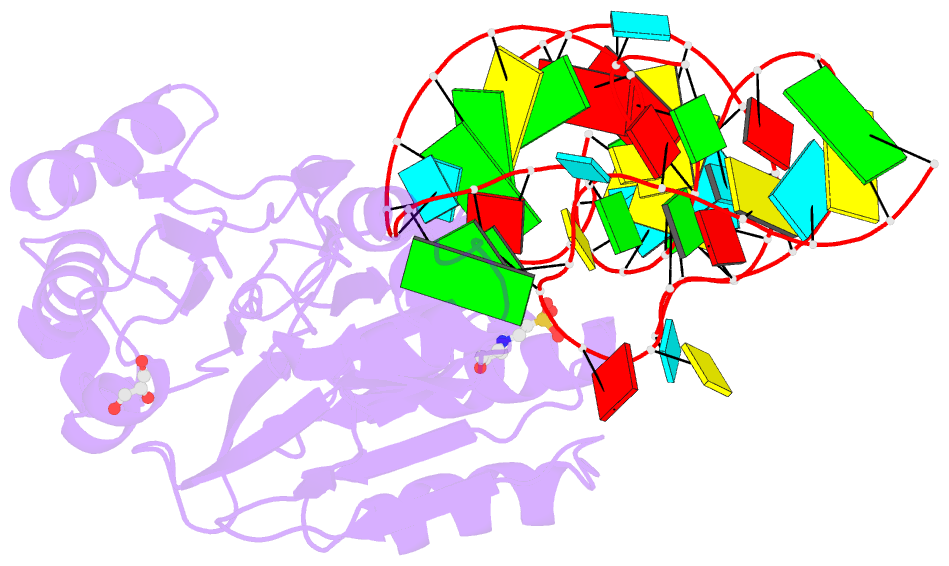
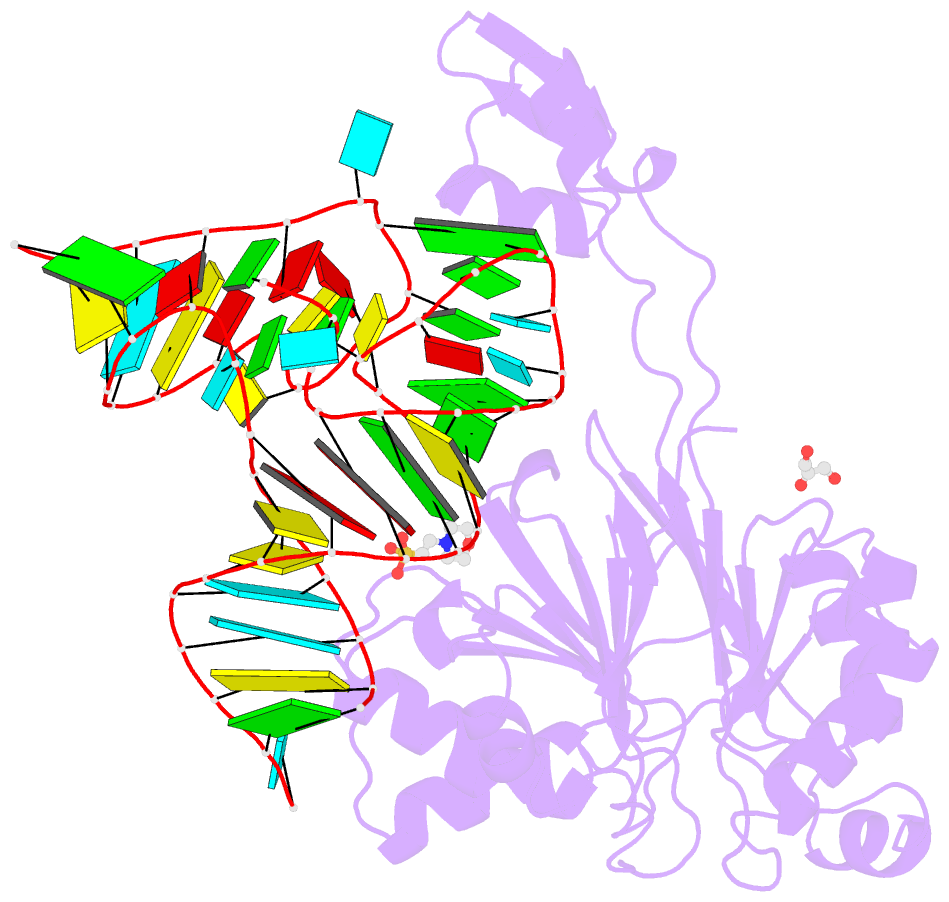
- Summary
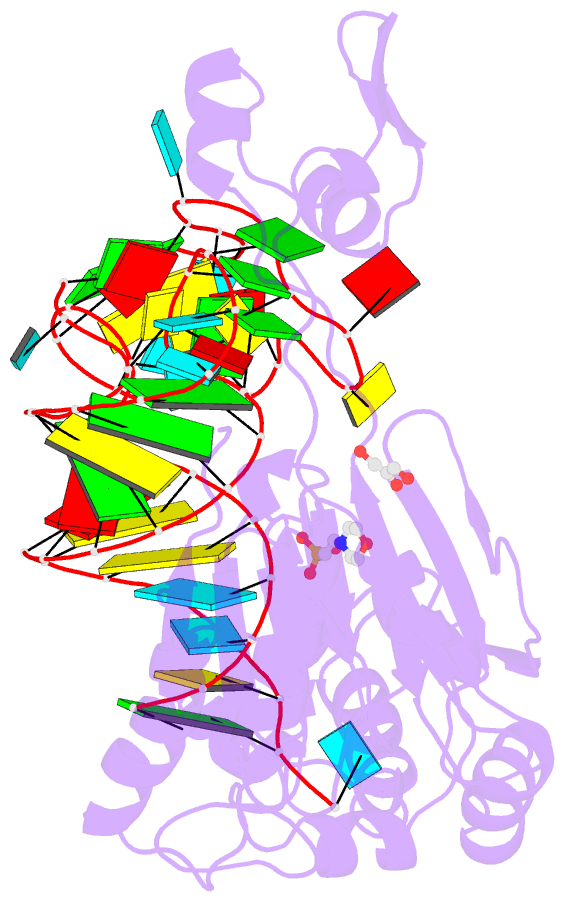
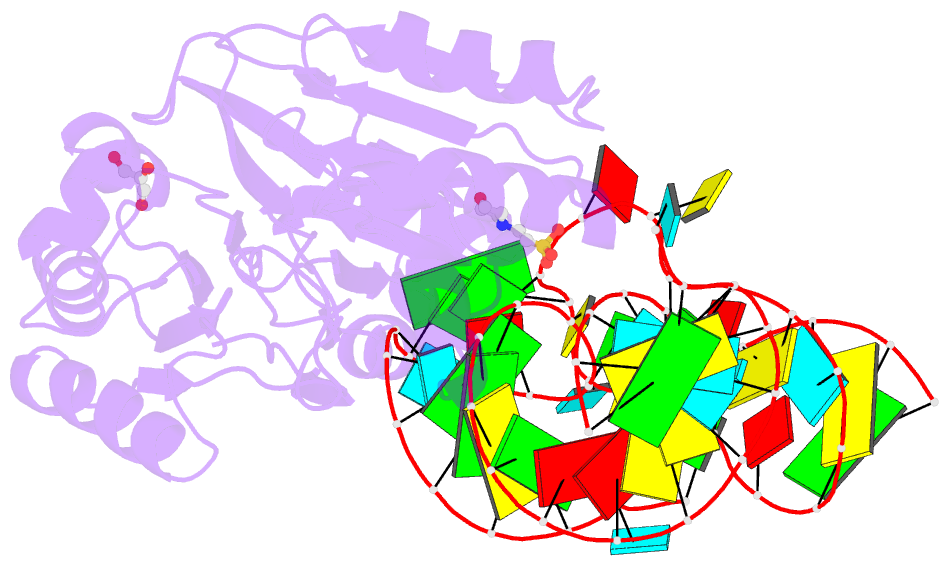
- Crystal structure of rnase z-trna(thr) complex
- Reference
- Li de la Sierra-Gallay I, Mathy N, Pellegrini O, Condon C (2006): "Structure of the ubiquitous 3' processing enzyme RNase Z bound to transfer RNA." Nat.Struct.Mol.Biol., 13, 376-377. doi: 10.1038/nsmb1066.
- Abstract
- The highly conserved ribonuclease RNase Z catalyzes the endonucleolytic removal of the 3' extension of the majority of tRNA precursors. Here we present the structure of the complex between Bacillus subtilis RNase Z and tRNA(Thr), the first structure of a ribonucleolytic processing enzyme bound to tRNA. Binding of tRNA to RNase Z causes conformational changes in both partners to promote reorganization of the catalytic site and tRNA cleavage.