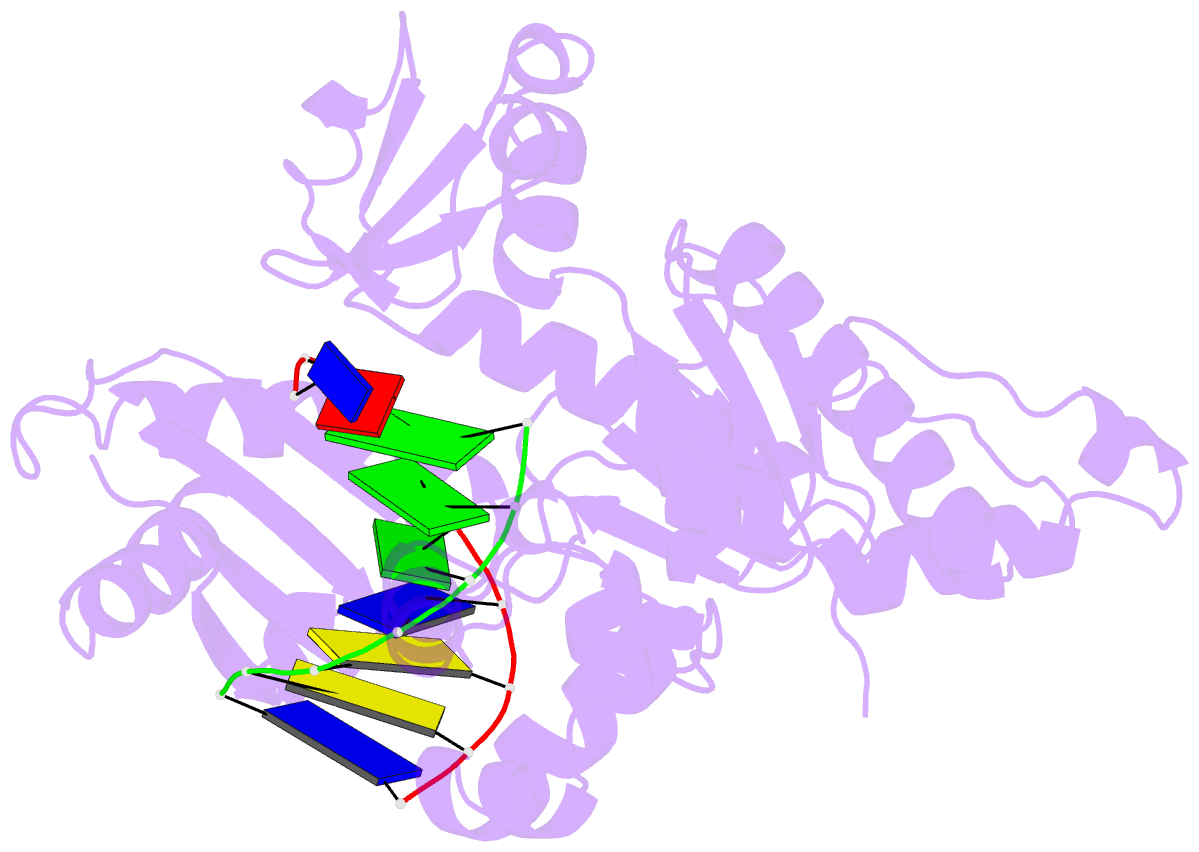
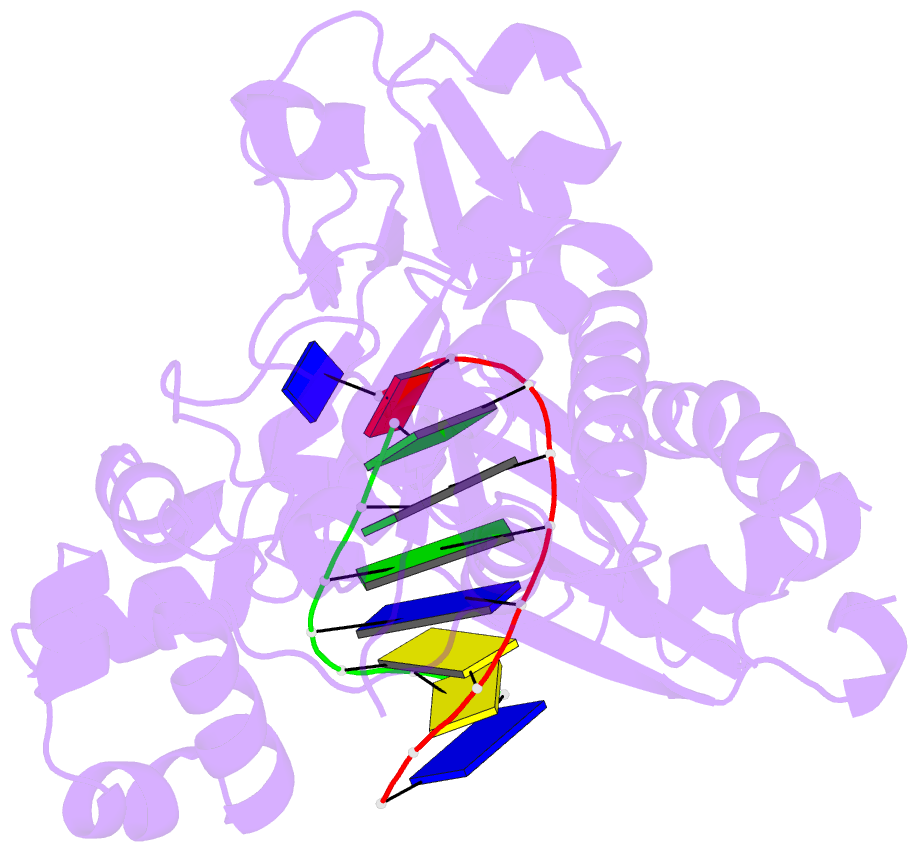
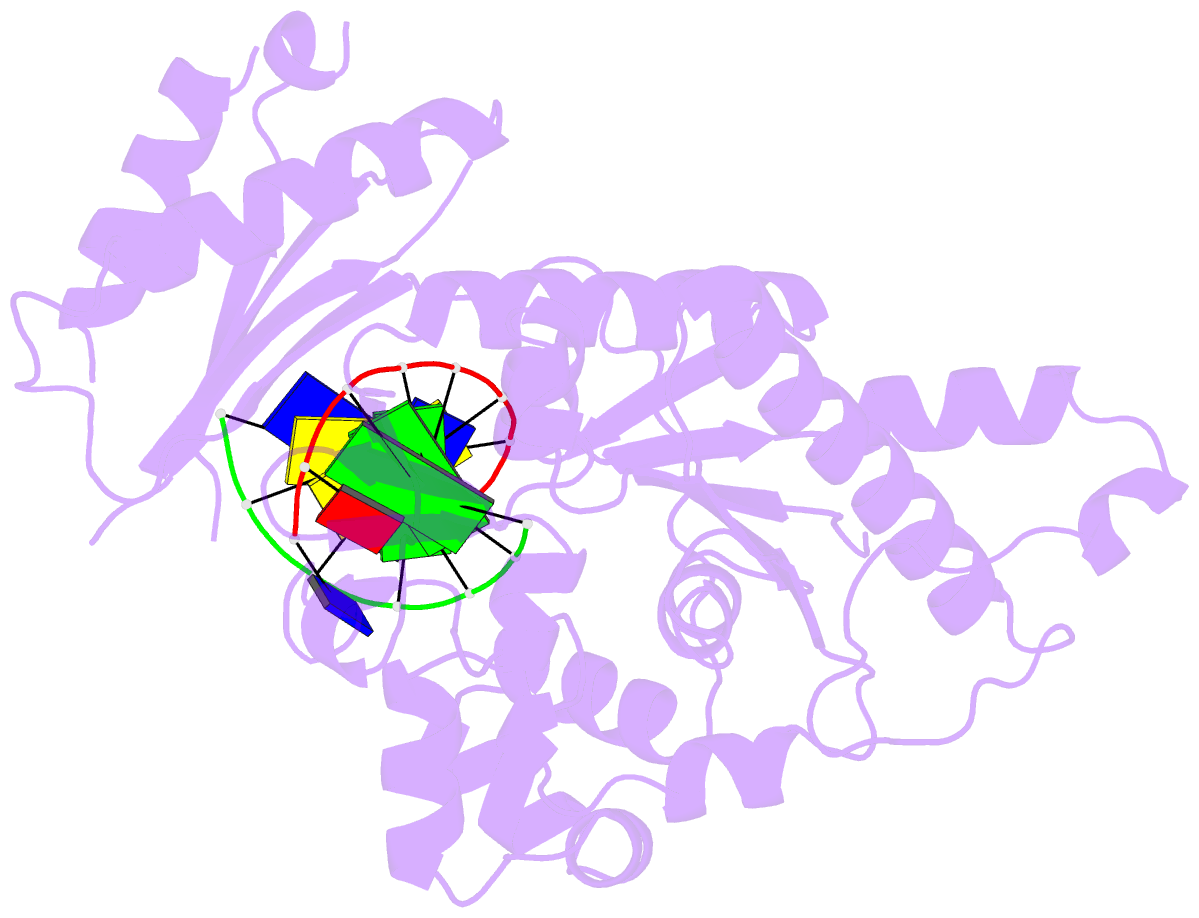
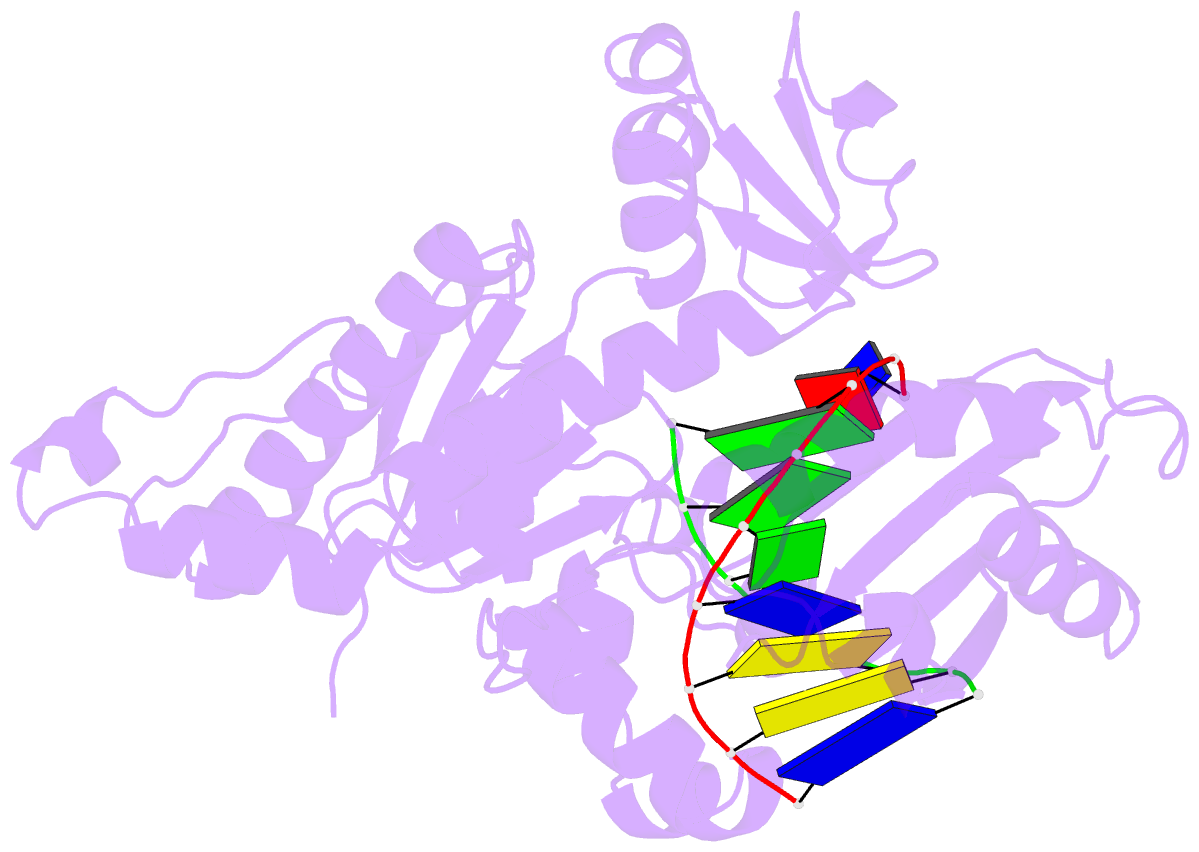
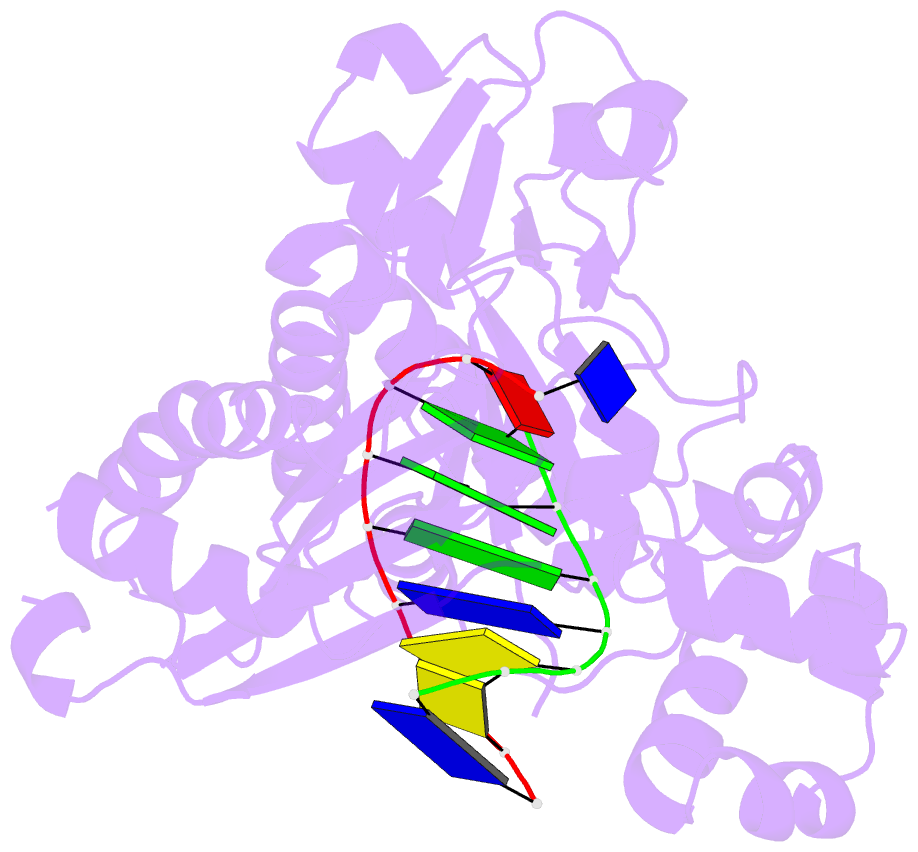
Summary information and primary citation
- PDB-id
- 2fln; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication-DNA
- Method
- X-ray (2.5 Å)
- Summary
- Binary complex of catalytic core of human DNA polymerase iota with DNA (template a)
- Reference
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK (2006): "An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-iota active site." Structure, 14, 749-755. doi: 10.1016/j.str.2006.01.010.
- Abstract
- Substrate-induced conformational change of the protein is the linchpin of enzymatic reactions. Replicative DNA polymerases, for example, convert from an open to a closed conformation in response to dNTP binding. Human DNA polymerase-iota (hPoliota), a member of the Y family of DNA polymerases, differs strikingly from other polymerases in its much higher proficiency and fidelity for nucleotide incorporation opposite template purines than opposite template pyrimidines. We present here a crystallographic analysis of hPoliota binary complexes, which together with the ternary complexes show that, contrary to replicative DNA polymerases, the DNA, and not the polymerase, undergoes the primary substrate-induced conformational change. The incoming dNTP "pushes" templates A and G from the anti to the syn conformation dictated by a rigid hPoliota active site. Together, the structures posit a mechanism for template selection wherein dNTP binding induces a conformational switch in template purines for productive Hoogsteen base pairing.