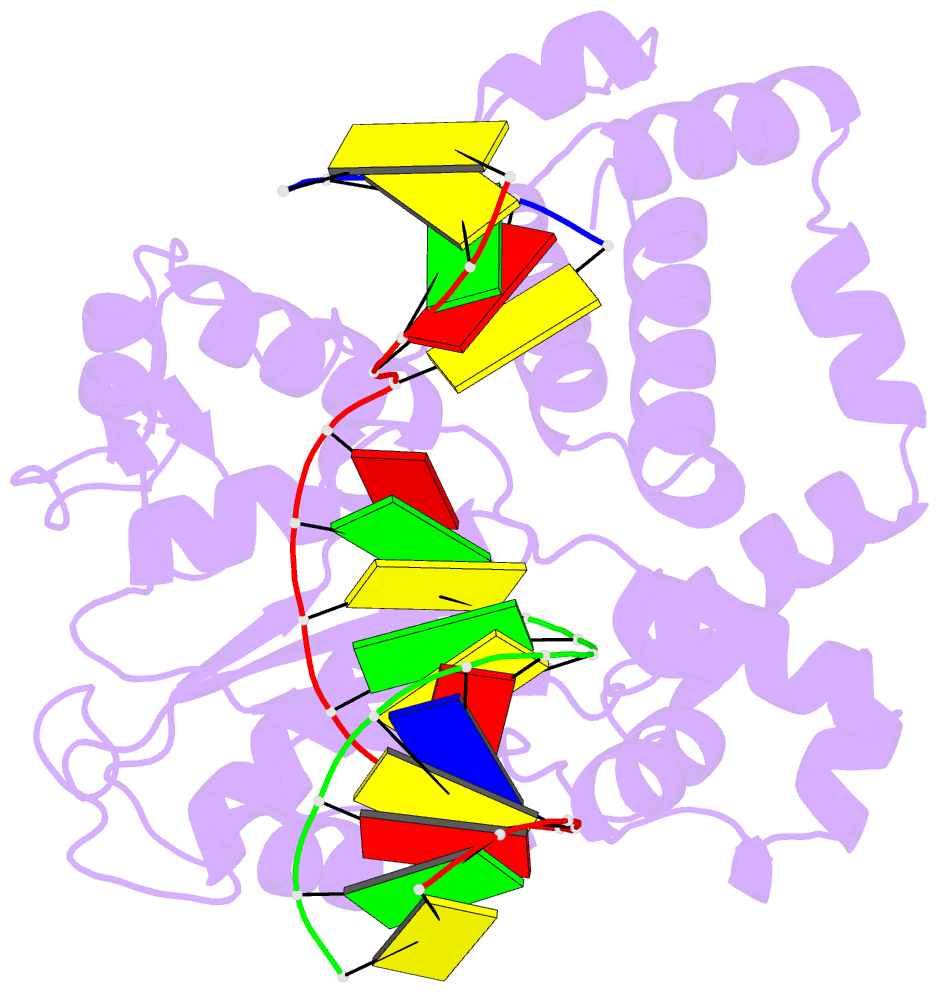
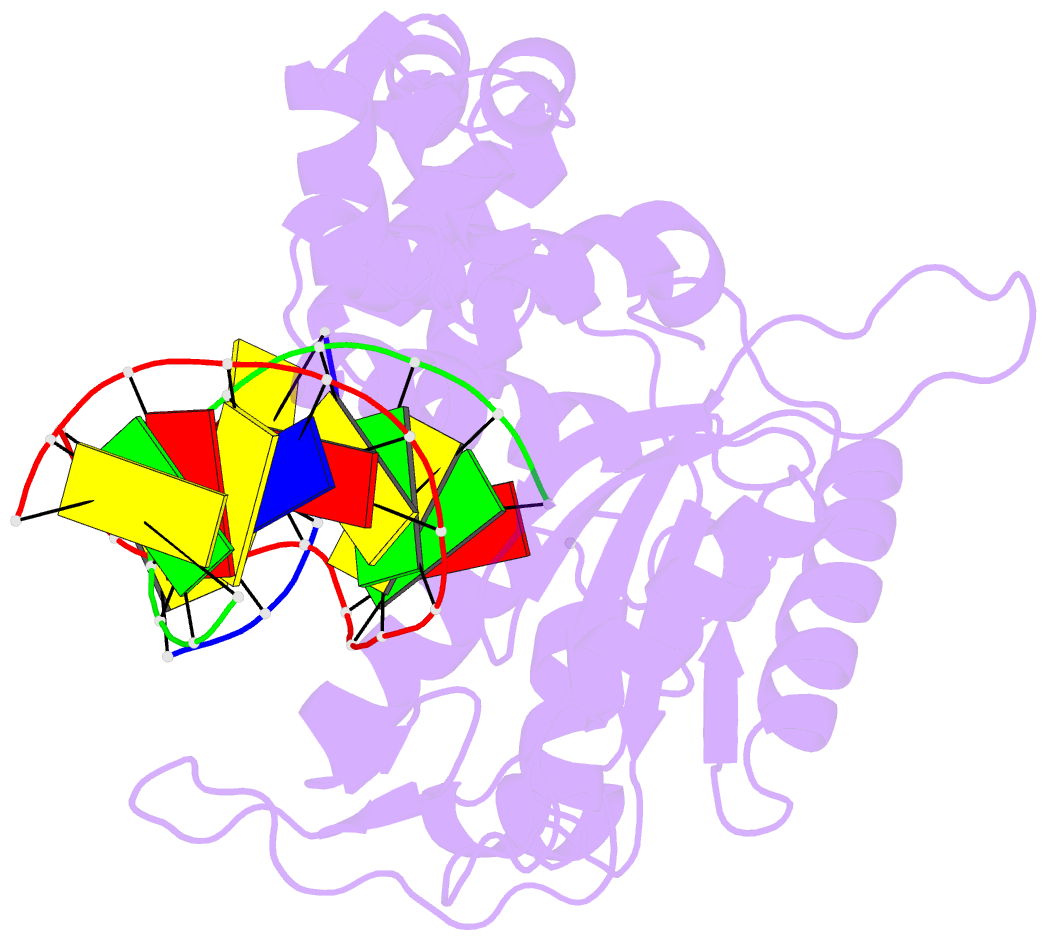
Summary information and primary citation
- PDB-id
- 2fmq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.2 Å)
- Summary
- Sodium in active site of DNA polymerase beta
- Reference
- Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC, Wilson SH (2006): "Magnesium-induced assembly of a complete DNA polymerase catalytic complex." Structure, 14, 757-766. doi: 10.1016/j.str.2006.01.011.
- Abstract
- The molecular details of the nucleotidyl transferase reaction have remained speculative, as strategies to trap catalytic intermediates for structure determination utilize substrates lacking the primer terminus 3'-OH and catalytic Mg2+, resulting in an incomplete and distorted active site geometry. Since the geometric arrangement of these essential atoms will impact chemistry, structural insight into fidelity strategies has been hampered. Here, we present a crystal structure of a precatalytic complex of a DNA polymerase with bound substrates that include the primer 3'-OH and catalytic Mg2+. This catalytic intermediate was trapped with a nonhydrolyzable deoxynucleotide analog. Comparison with two new structures of DNA polymerase beta lacking the 3'-OH or catalytic Mg2+ is described. These structures provide direct evidence that both atoms are required to achieve a proper geometry necessary for an in-line nucleophilic attack of O3' on the alphaP of the incoming nucleotide.