Summary information and primary citation
- PDB-id
- 2fmt; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- complex (methyltransferase-trna)
- Method
- X-ray (2.8 Å)
- Summary
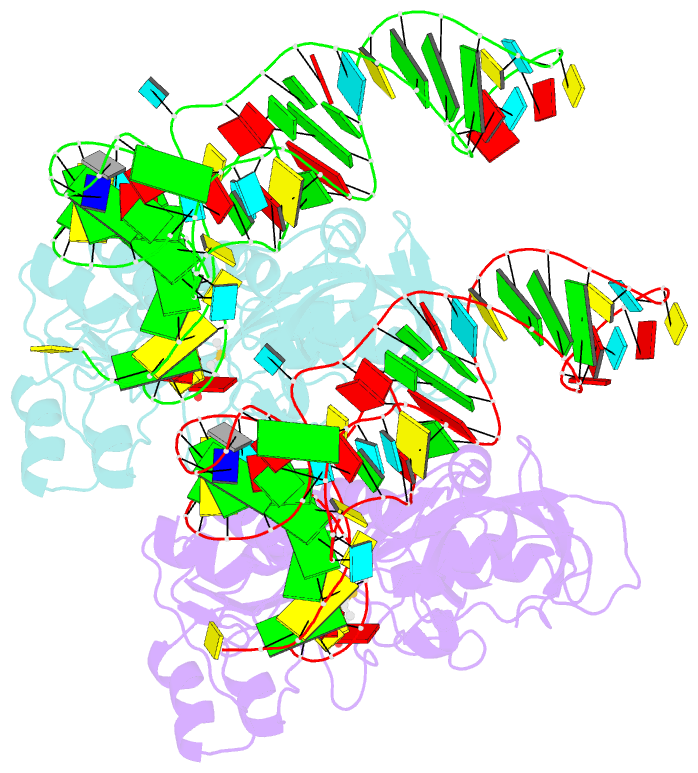
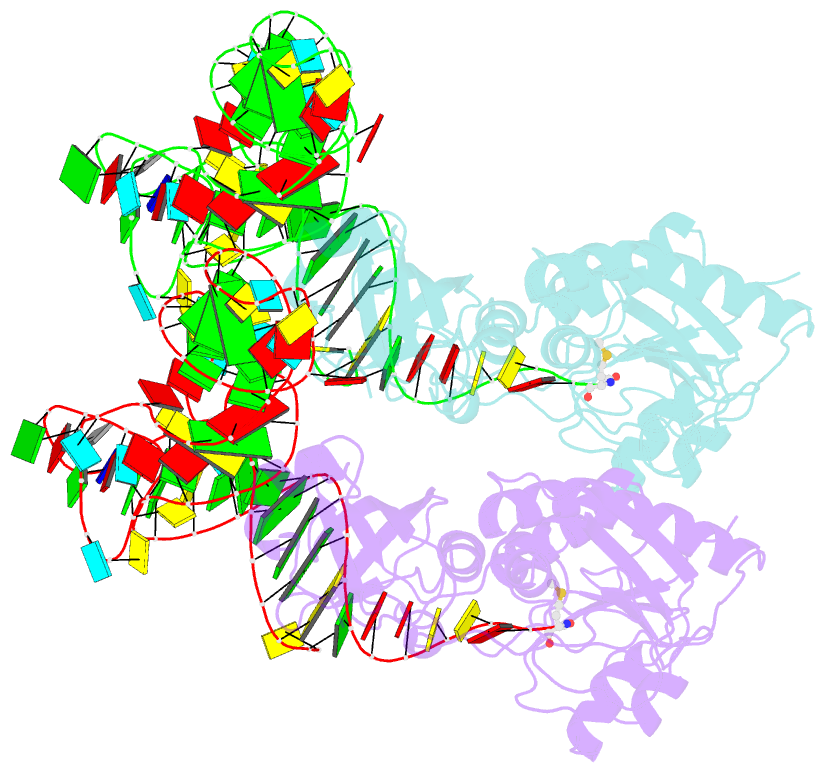
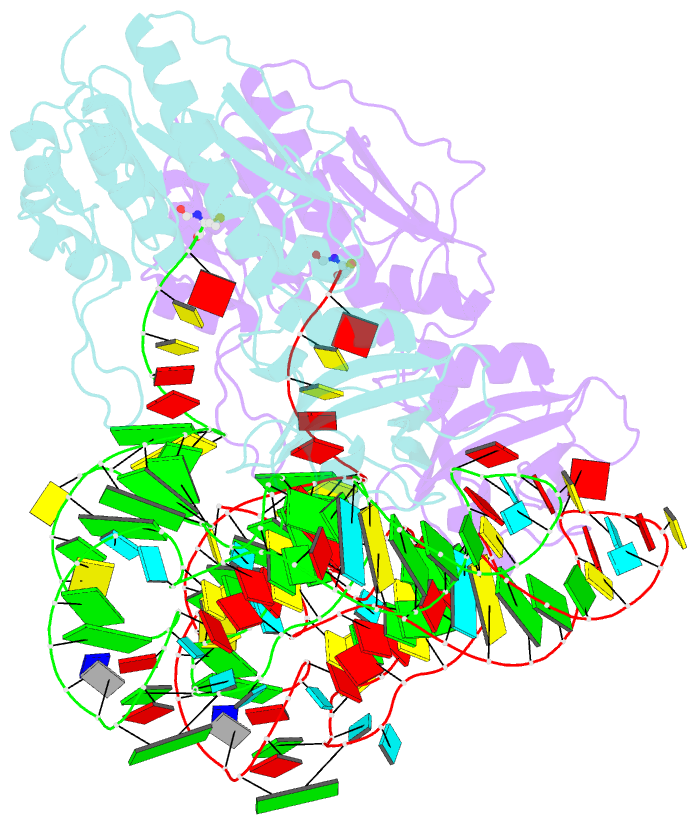
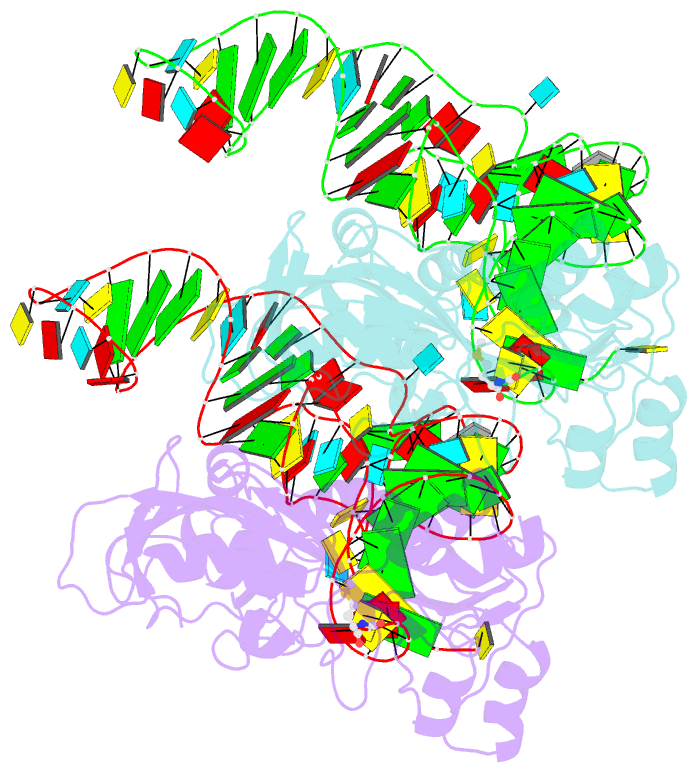
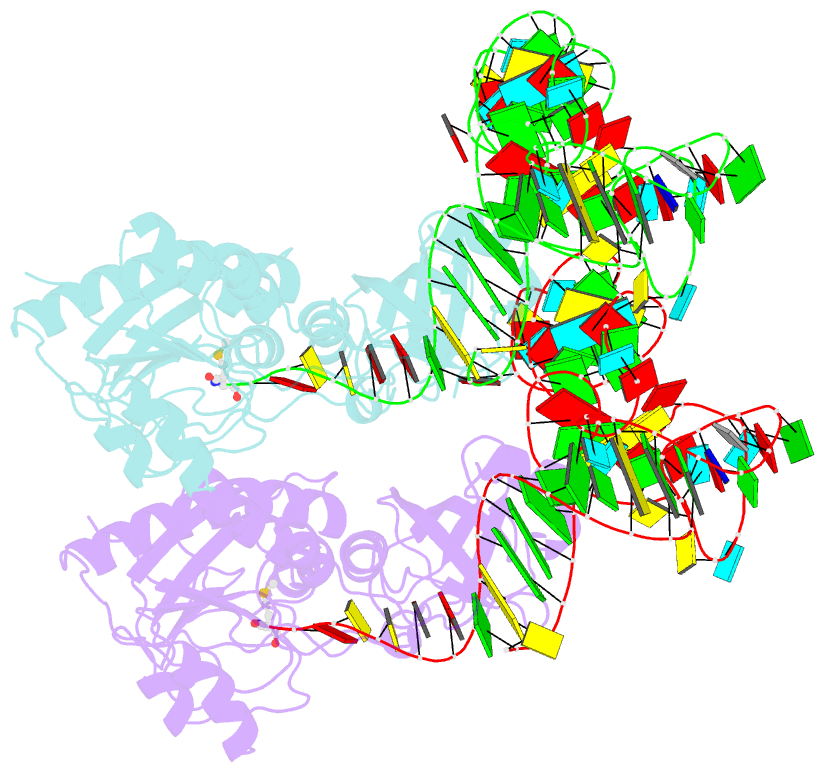
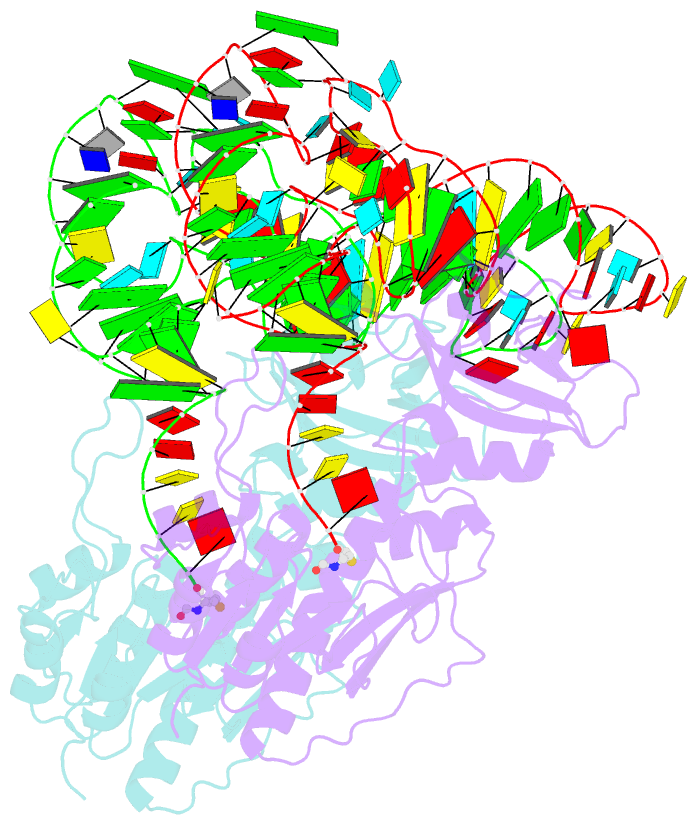
- Methionyl-trnafmet formyltransferase complexed with formyl-methionyl-trnafmet
- Reference
- Schmitt E, Panvert M, Blanquet S, Mechulam Y (1998): "Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet." EMBO J., 17, 6819-6826. doi: 10.1093/emboj/17.23.6819.
- Abstract
- The crystal structure of Escherichia coli methionyl-tRNAfMet transformylase complexed with formyl-methionyl-tRNAfMet was solved at 2.8 A resolution. The formylation reaction catalyzed by this enzyme irreversibly commits methionyl-tRNAfMet to initiation of translation in eubacteria. In the three-dimensional model, the methionyl-tRNAfMet formyltransferase fills in the inside of the L-shaped tRNA molecule on the D-stem side. The anticodon stem and loop are away from the protein. An enzyme loop is wedged in the major groove of the acceptor helix. As a result, the C1-A72 mismatch characteristic of the initiator tRNA is split and the 3' arm bends inside the active centre. This recognition mechanism is markedly distinct from that of elongation factor Tu, which binds the acceptor arm of aminoacylated elongator tRNAs on the T-stem side.