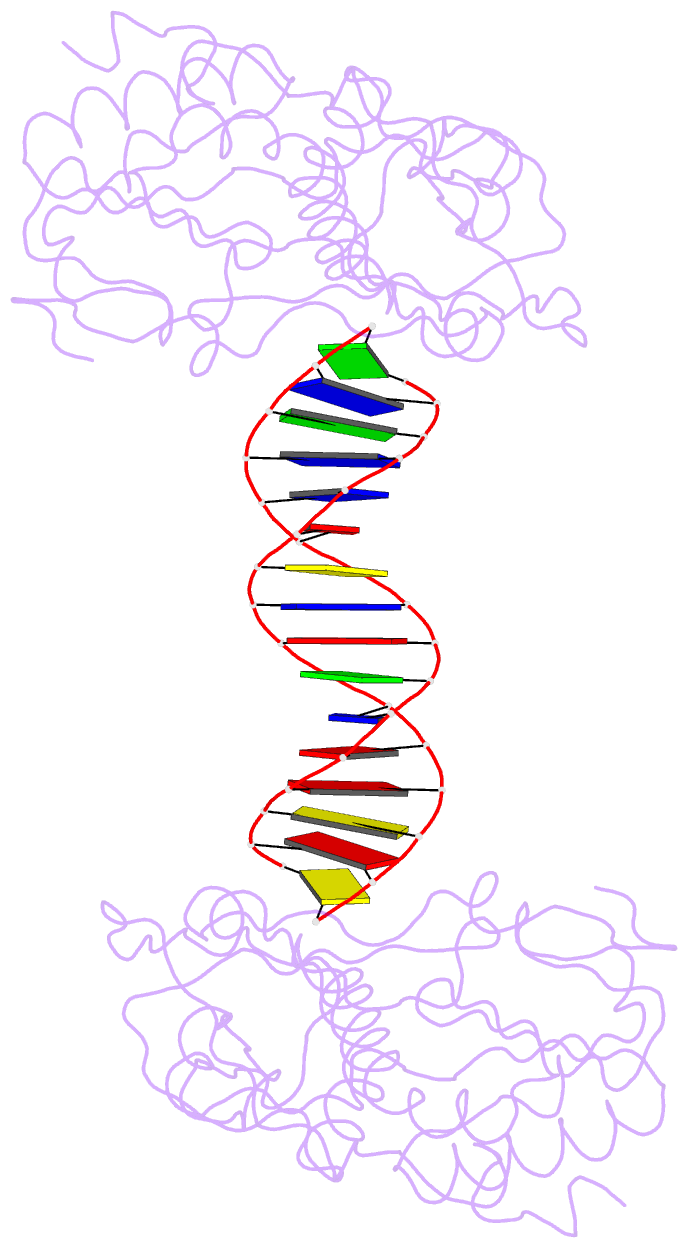
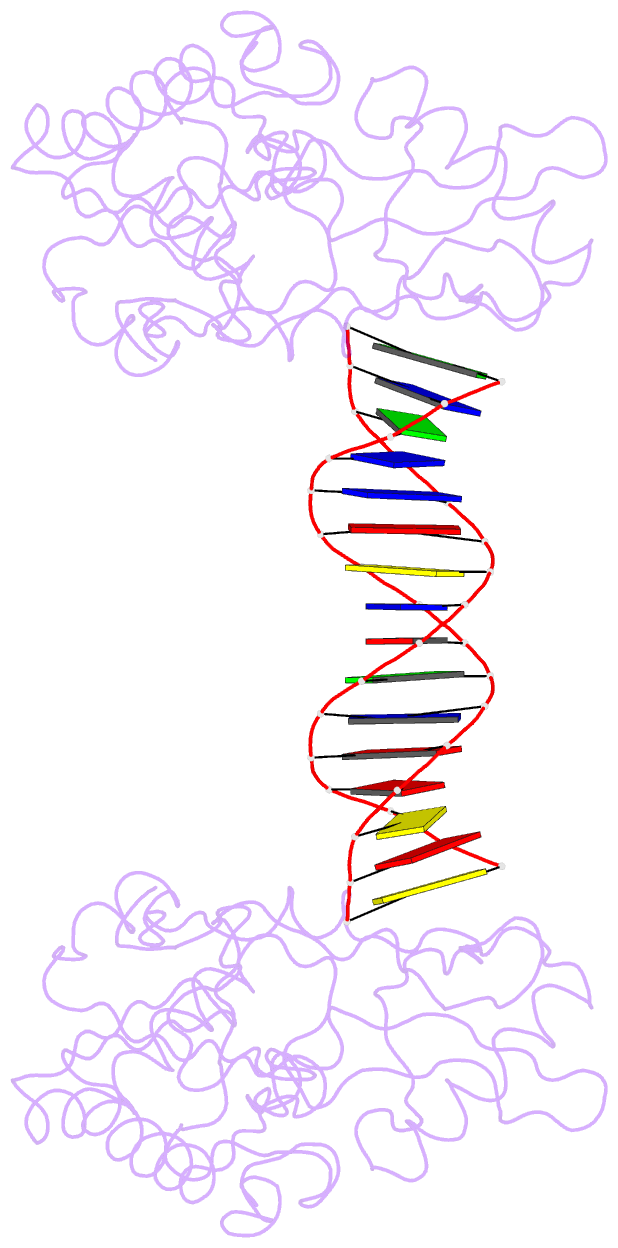
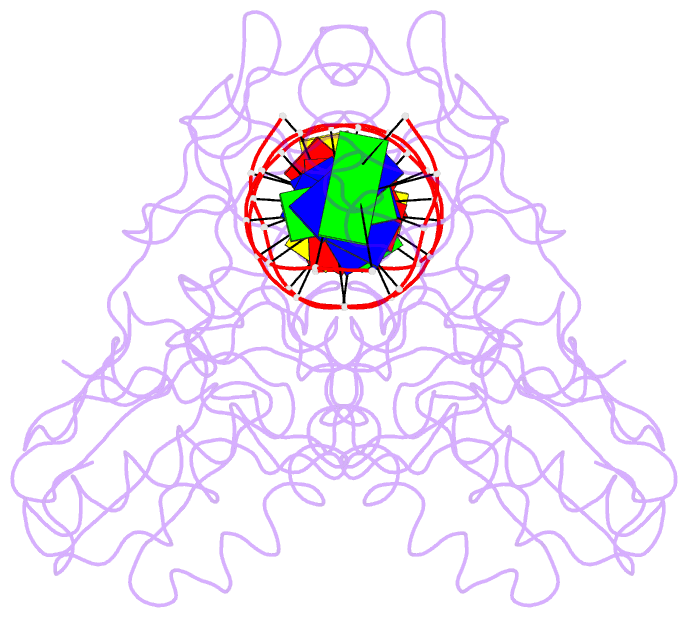
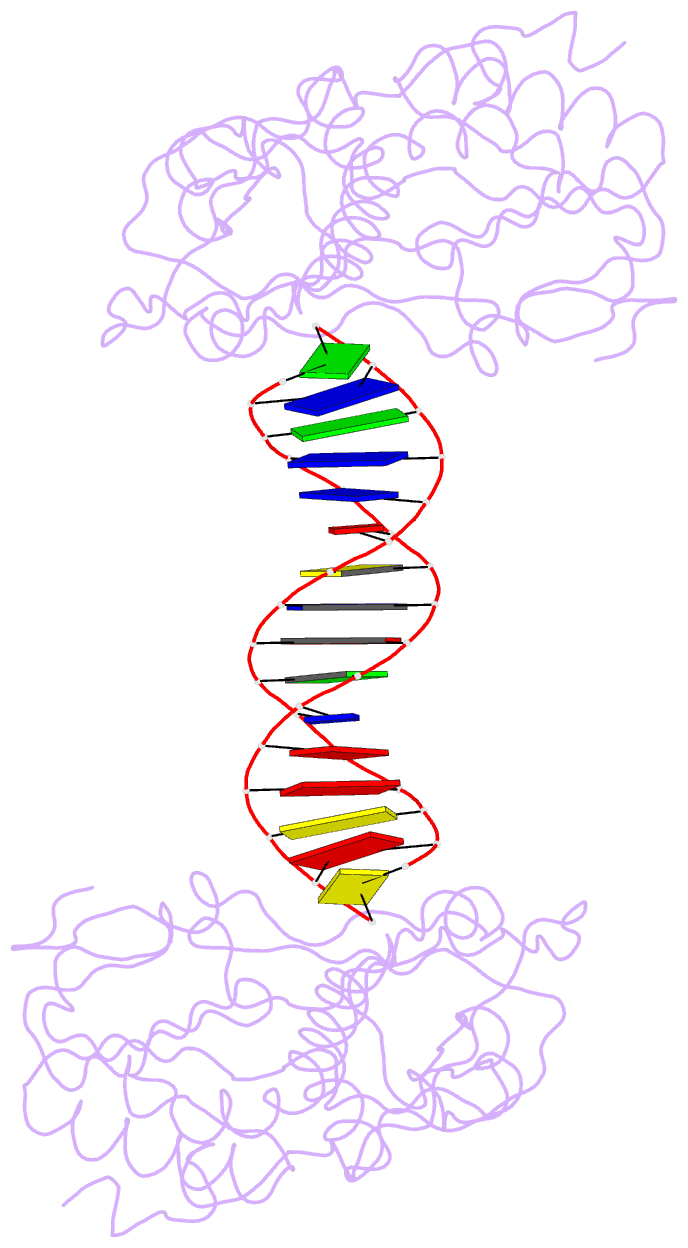
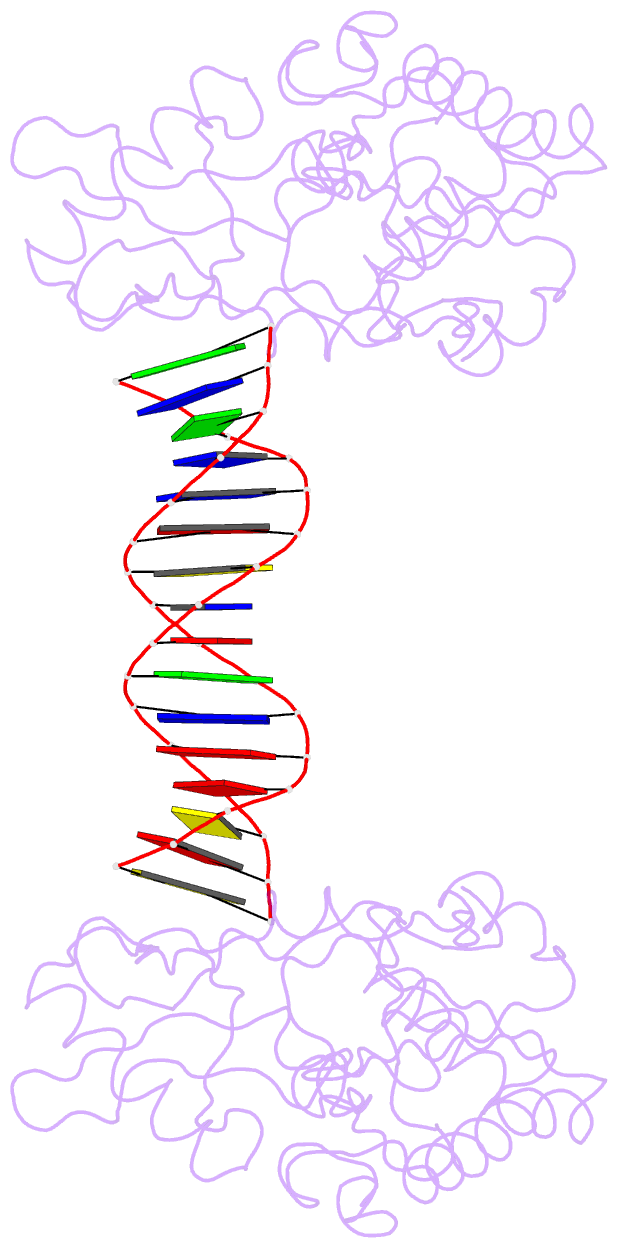
Summary information and primary citation
- PDB-id
- 2fvs; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.35 Å)
- Summary
- A structural study of the ca dinucleotide step in the integrase processing site of moloney murine leukemia virus
- Reference
- Montano SP, Cote ML, Roth MJ, Georgiadis MM (2006): "Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus." Nucleic Acids Res., 34, 5353-5360. doi: 10.1093/nar/gkl693.
- Abstract
- In the first step of retroviral integration, integrase cleaves the linear viral DNA within its long terminal repeat (LTR) immediately 3' to the CA dinucleotide step, resulting in a reactive 3' OH on one strand and a 5' two base overhang on the complementary strand. In order to investigate the structural properties of the 3' end processing site within the Moloney murine leukemia virus (MMLV) LTR d(TCTTTCATT), a host-guest crystallographic method was employed to determine the structures of four self-complementary 16 bp oligonucleotides including LTR sequences (underlined), d(TTTCATTGCAATGAAA), d(CTTTCATTAATGAAAG), d(TCTTTCATATGAAAGA) and d(CACAATGATCATTGTG), the guests, complexed with the N-terminal fragment of MMLV reverse transcriptase, the host. The structures of the LTR-containing oligonucleotides were compared to those of non-LTR oligonucleotides crystallized in the same lattice. Properties unique to the CA dinucleotide step within the LTR sequence, independent of its position from the end of the duplex, include a positive roll angle and negative slide value. This propensity for the CA dinucleotide step within the MMLV LTR sequence to adopt only positive roll angles is likely influenced by the more rigid, invariable 3' and 5' flanking TT dinucleotide steps and may be important for specific recognition and/or cleavage by the MMLV integrase.