Summary information and primary citation
- PDB-id
- 2g1p; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (1.89 Å)
- Summary
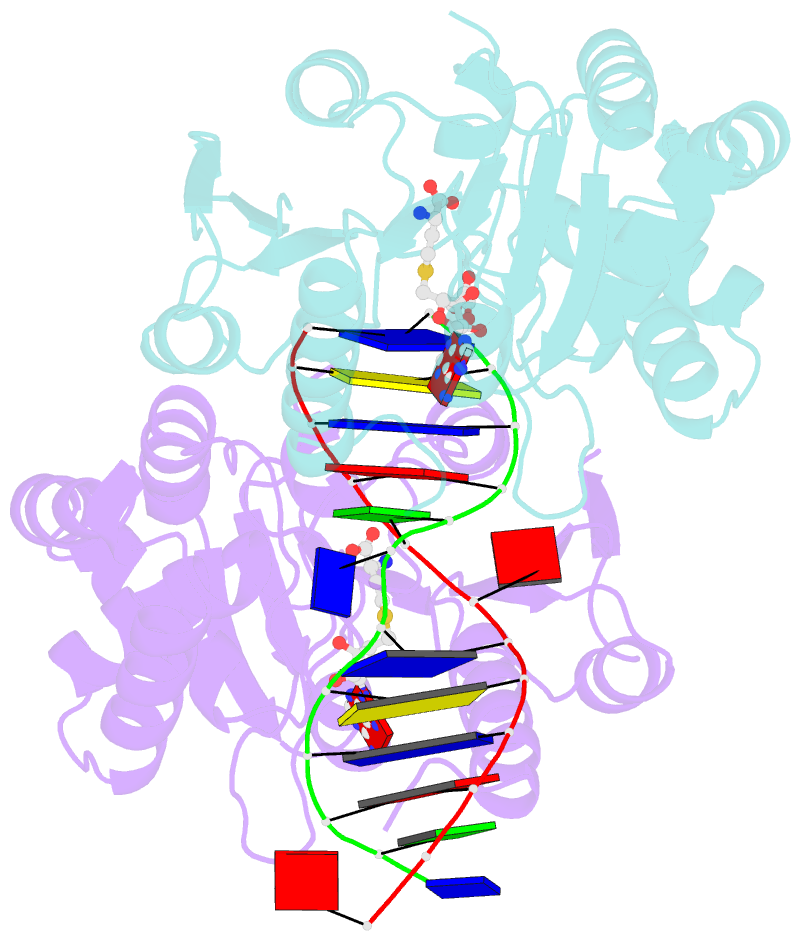
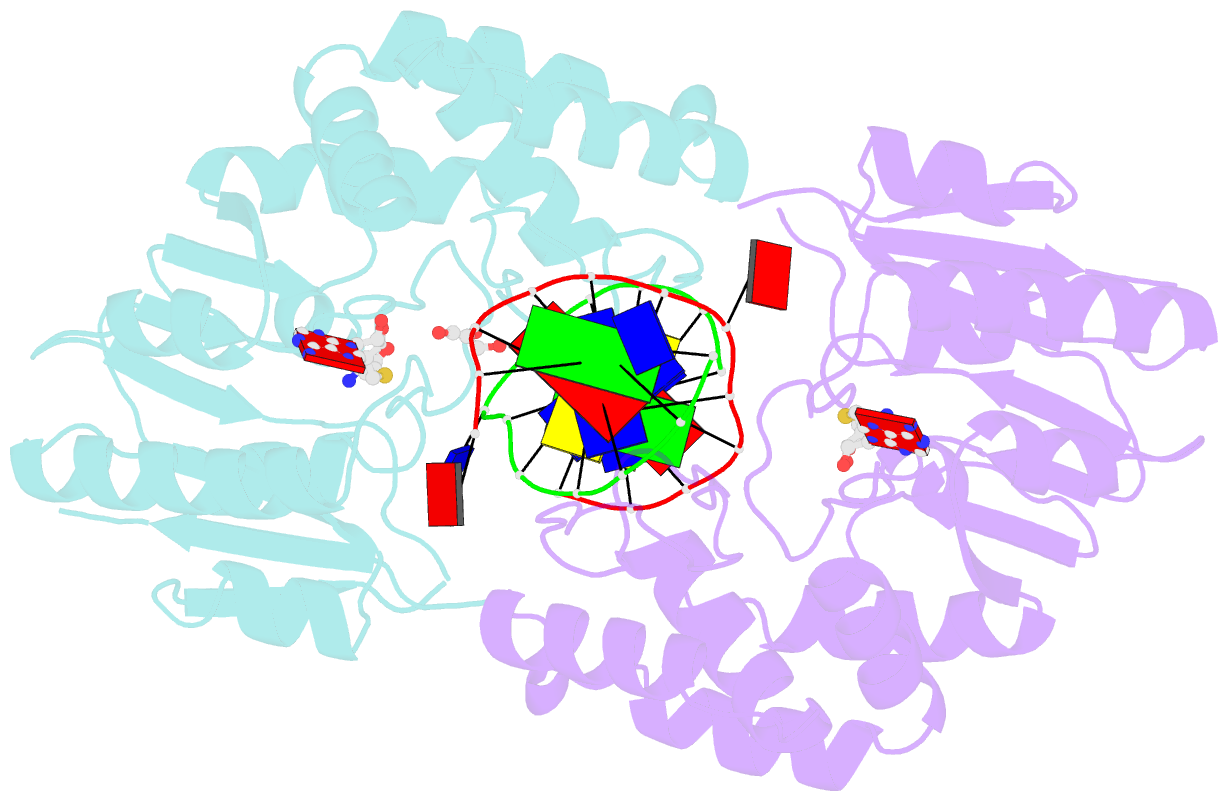
- Structure of e. coli DNA adenine methyltransferase (dam)
- Reference
- Horton JR, Liebert K, Bekes M, Jeltsch A, Cheng X (2006): "Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase." J.Mol.Biol., 358, 559-570. doi: 10.1016/j.jmb.2006.02.028.
- Abstract
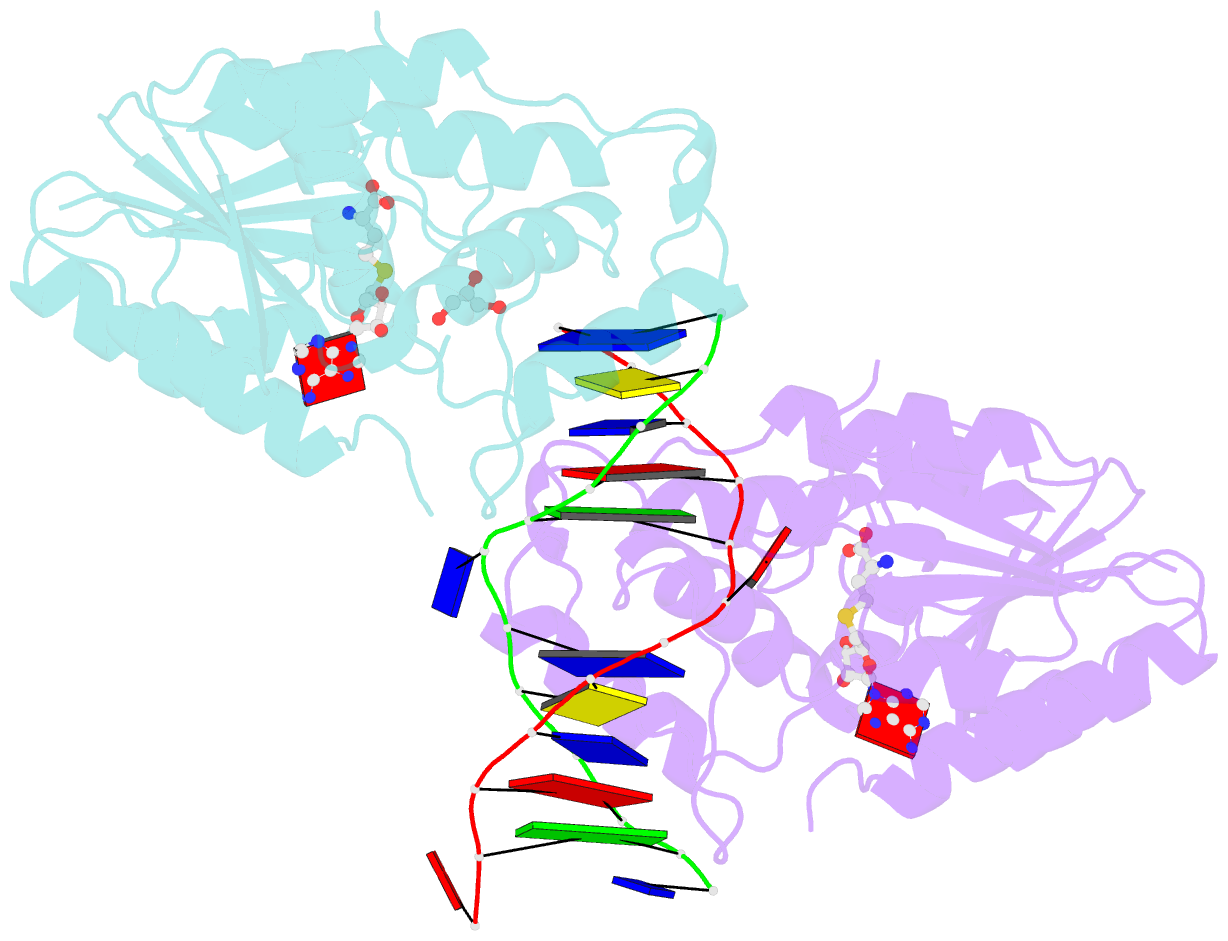
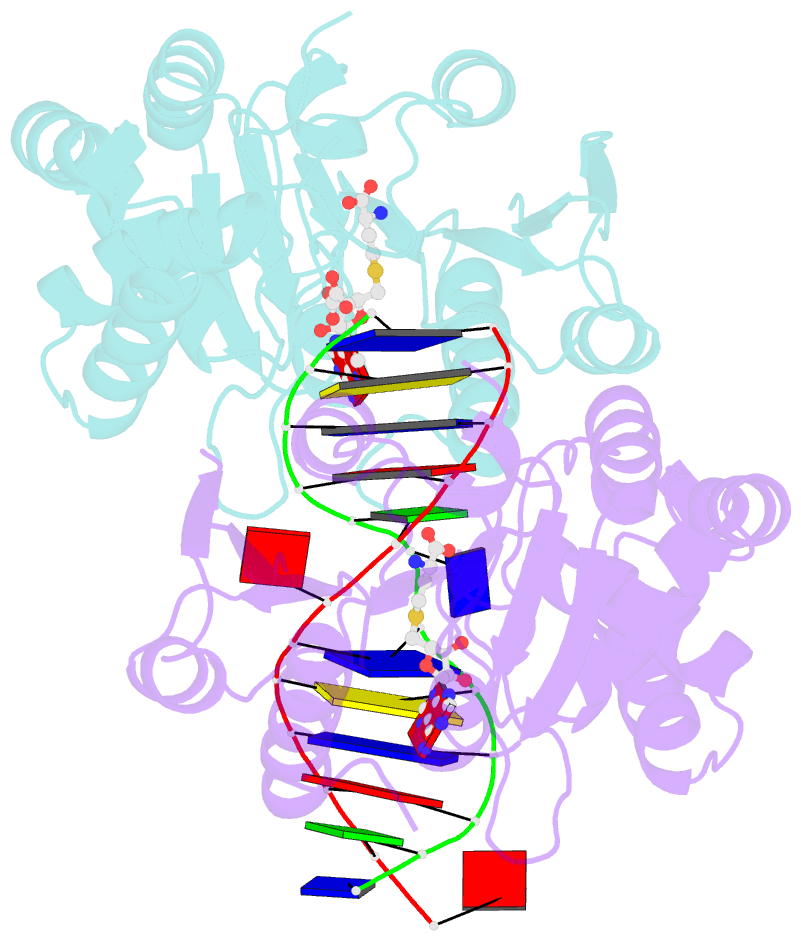
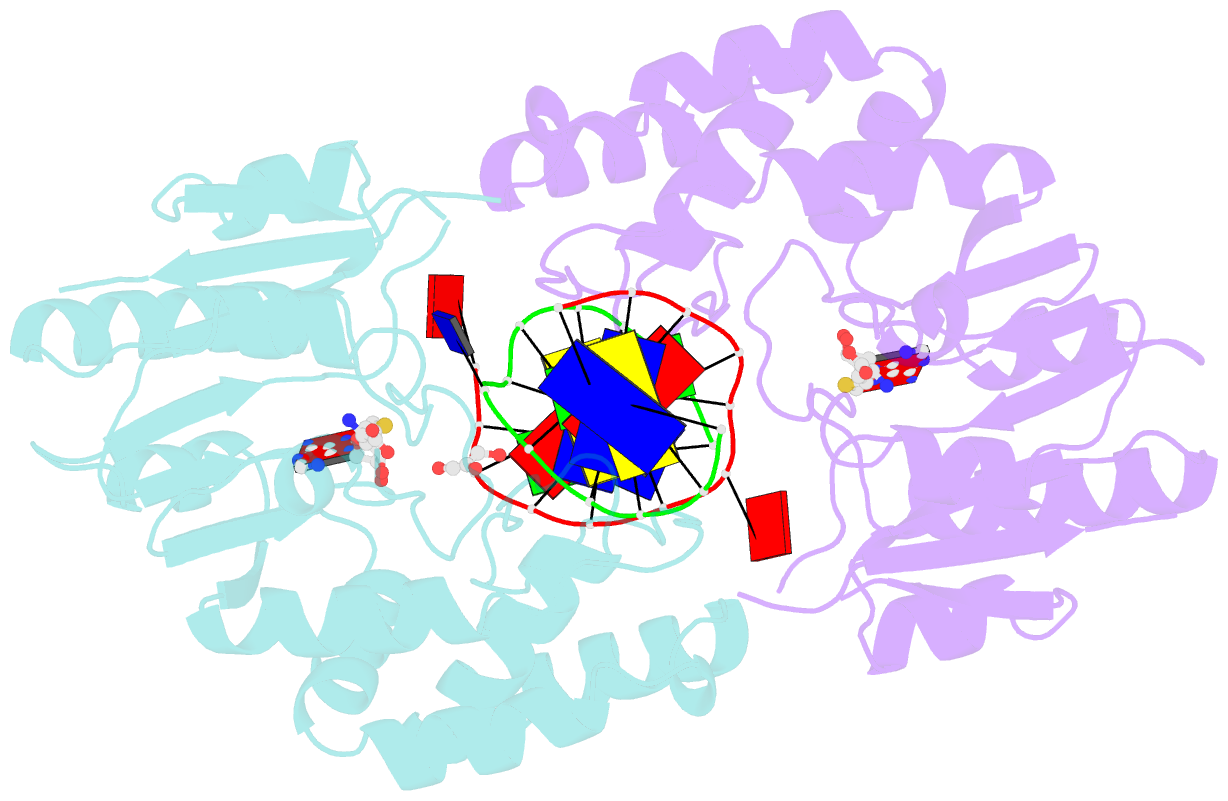
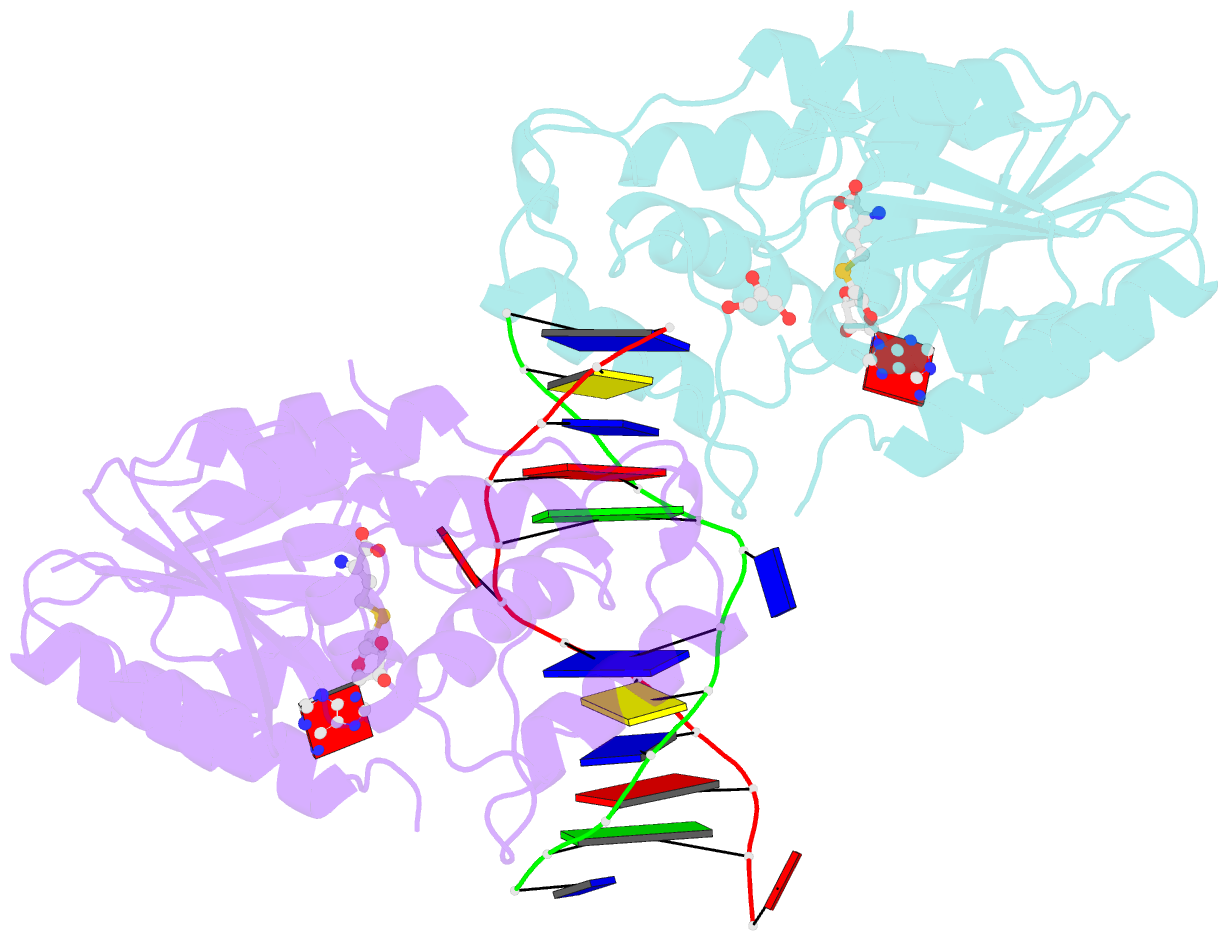
- The structure of the Escherichia coli Dam DNA-(adenine-N6)-methyltransferase in complex with cognate DNA was determined at 1.89 A resolution in the presence of S-adenosyl-L-homocysteine. DNA recognition and the dynamics of base-flipping were studied by site-directed mutagenesis, DNA methylation kinetics and fluorescence stopped-flow experiments. Our data illustrate the mechanism of coupling of DNA recognition and base-flipping. Contacts to the non-target strand in the second (3') half of the GATC site are established by R124 to the fourth base-pair, and by L122 and P134 to the third base-pair. The aromatic ring of Y119 intercalates into the DNA between the second and third base-pairs, which is essential for base-flipping to occur. Compared to previous published structures of bacteriophage T4 Dam, three major new observations are made in E.coli Dam. (1) The first Gua is recognized by K9, removal of which abrogates the first base-pair recognition. (2) The flipped target Ade binds to the surface of EcoDam in the absence of S-adenosyl-L-methionine, which illustrates a possible intermediate in the base-flipping pathway. (3) The orphaned Thy residue displays structural flexibility by adopting an extrahelical or intrahelical position where it is in contact to N120.