Summary information and primary citation
- PDB-id
- 2geq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.3 Å)
- Summary
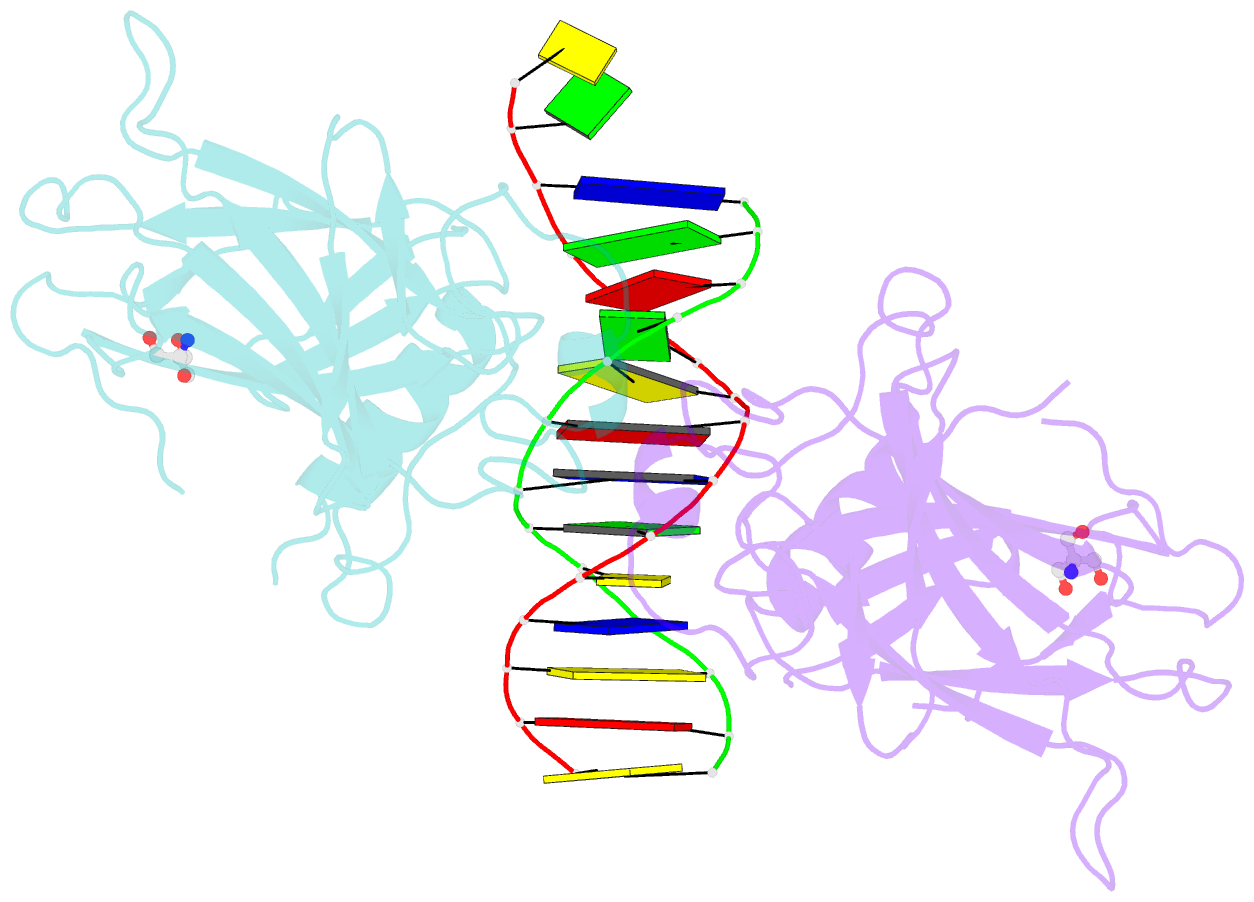
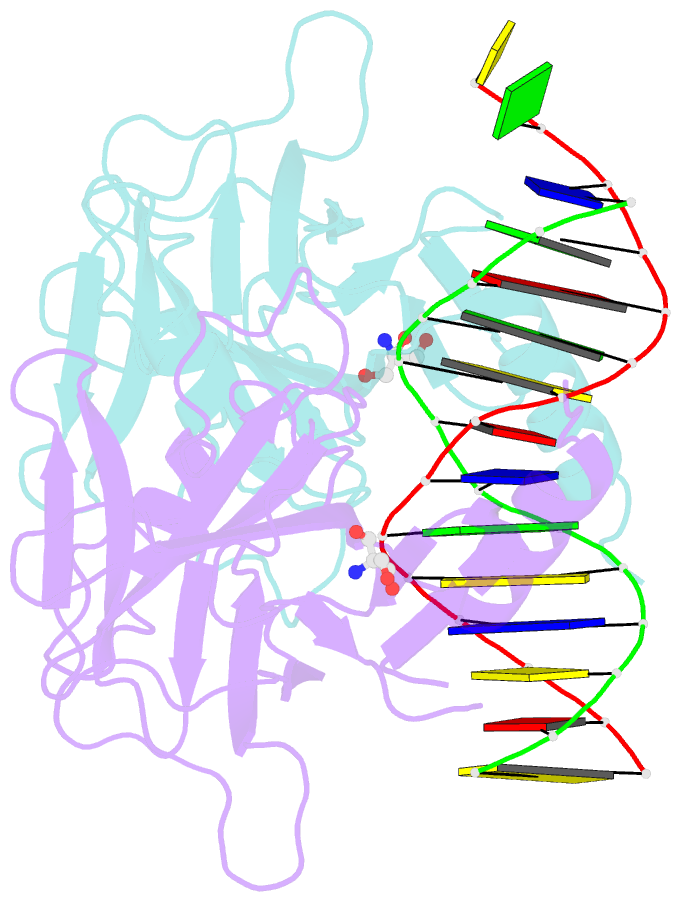
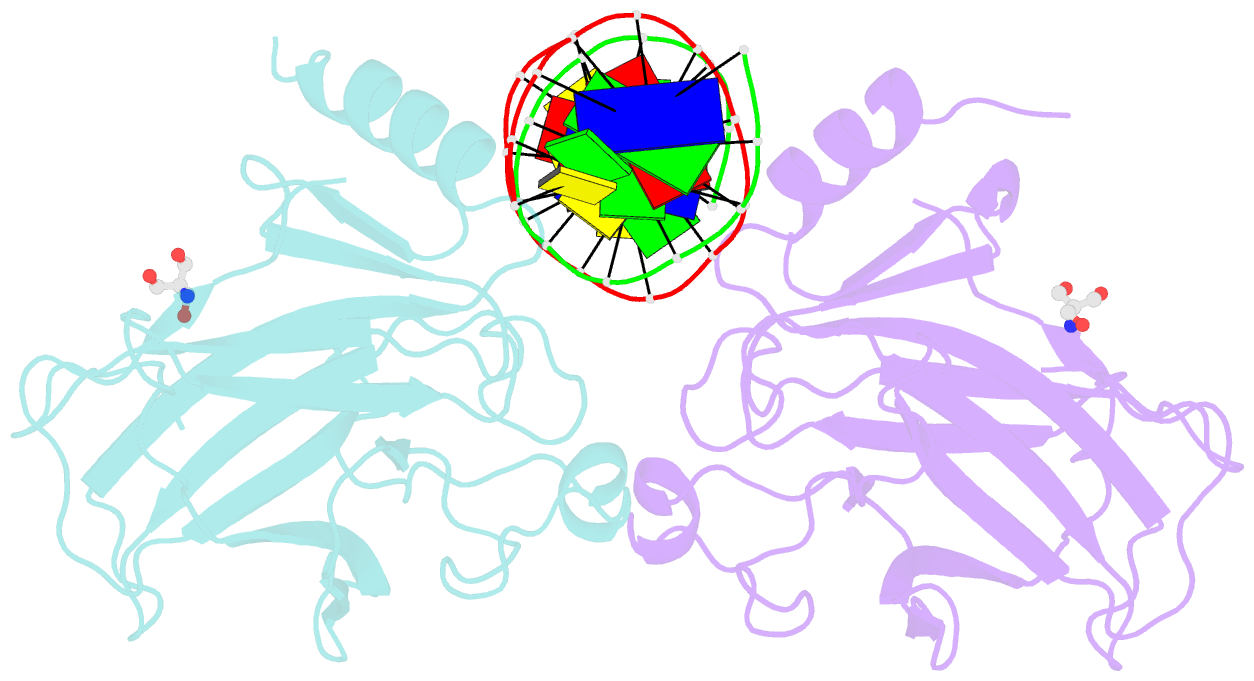
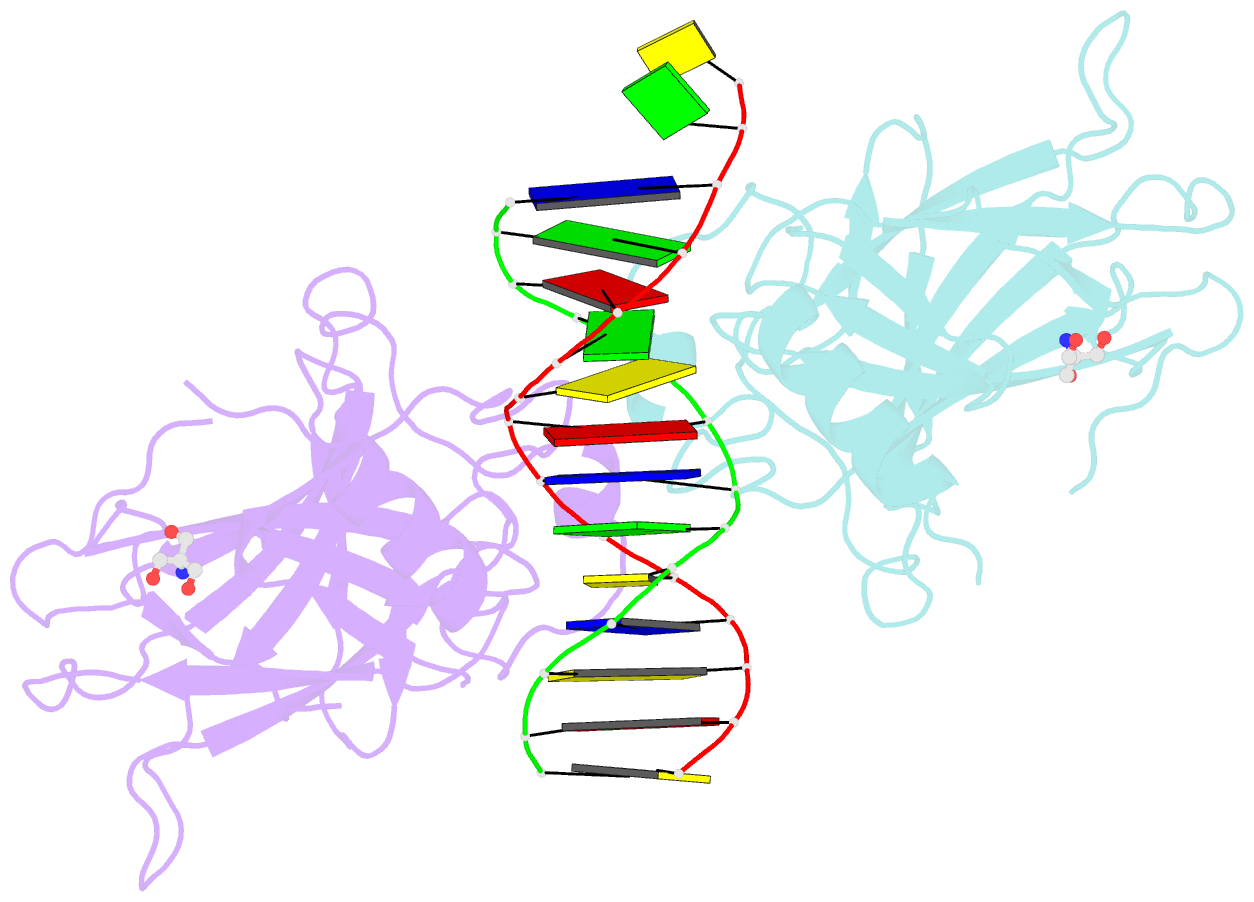
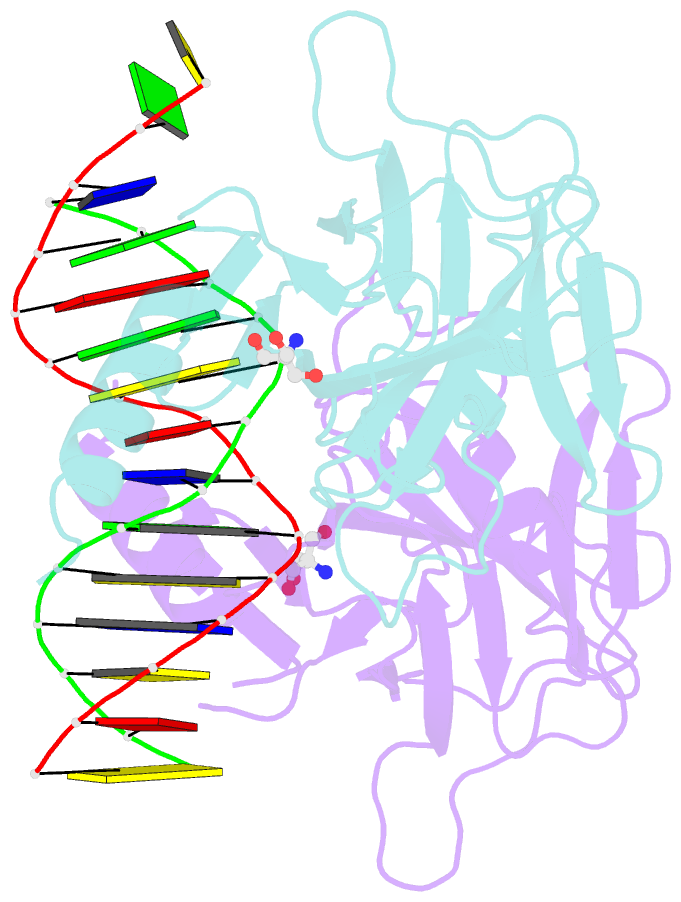
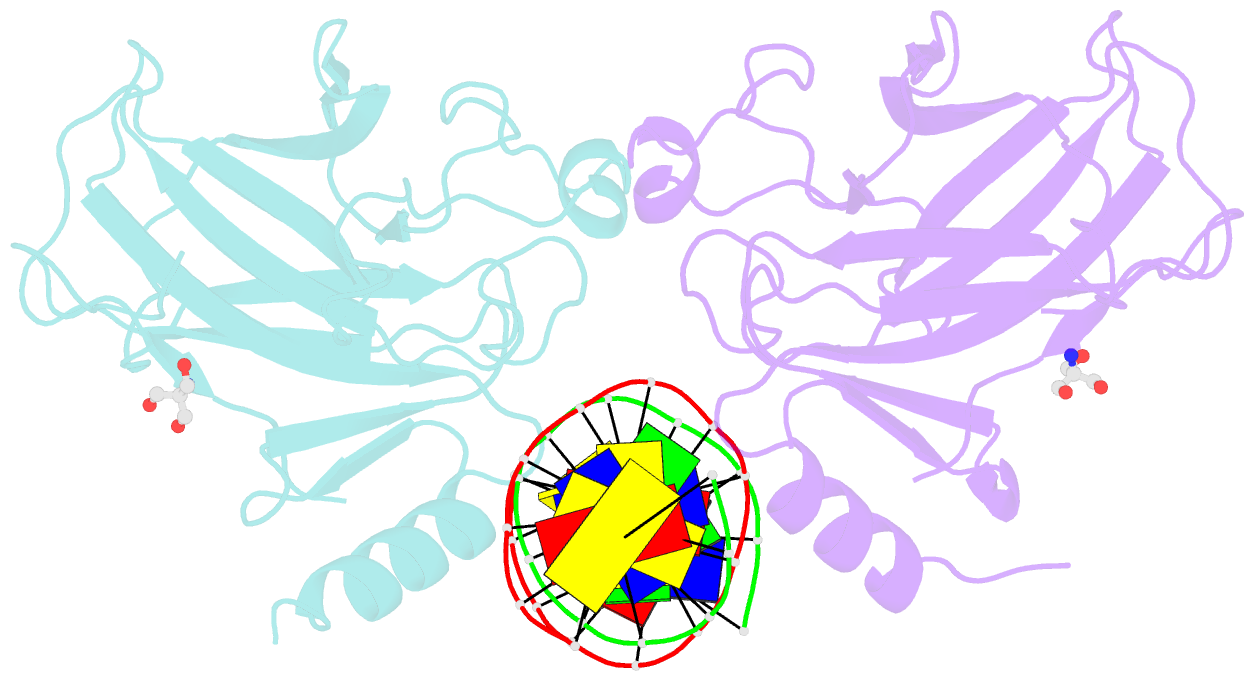
- Crystal structure of a p53 core dimer bound to DNA
- Reference
- Ho WC, Fitzgerald MX, Marmorstein R (2006): "Structure of the p53 Core Domain Dimer Bound to DNA." J.Biol.Chem., 281, 20494-20502. doi: 10.1074/jbc.M603634200.
- Abstract
- The p53 tumor suppressor protein binds to DNA as a dimer of dimers to regulate transcription of genes that mediate responses to cellular stress. We have prepared a cross-linked trapped p53 core domain dimer bound to decamer DNA and have determined its structure by x-ray crystallography to 2.3A resolution. The p53 core domain subunits bind nearly symmetrically to opposite faces of the DNA in a head-to-head fashion with a loophelix motif making sequence-specific DNA contacts and bending the DNA by about 20 degrees at the site of protein dimerization. Protein subunit interactions occur over the central DNA minor groove and involve residues from a zinc-binding region. Analysis of tumor derived p53 mutations reveals that the dimerization interface represents a third hot spot for mutation that also includes residues associated with DNA contact and protein stability. Residues associated with p53 dimer formation on DNA are poorly conserved in the p63 and p73 paralogs, possibly contributing to their functional differences. We have used the dimeric protein-DNA complex to model a dimer of p53 dimers bound to icosamer DNA that is consistent with solution bending data and suggests that p53 core domain dimer-dimer contacts are less frequently mutated in human cancer than intra-dimer contacts.