Summary information and primary citation
- PDB-id
- 2gje; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- translation-RNA
- Method
- X-ray (3.37 Å)
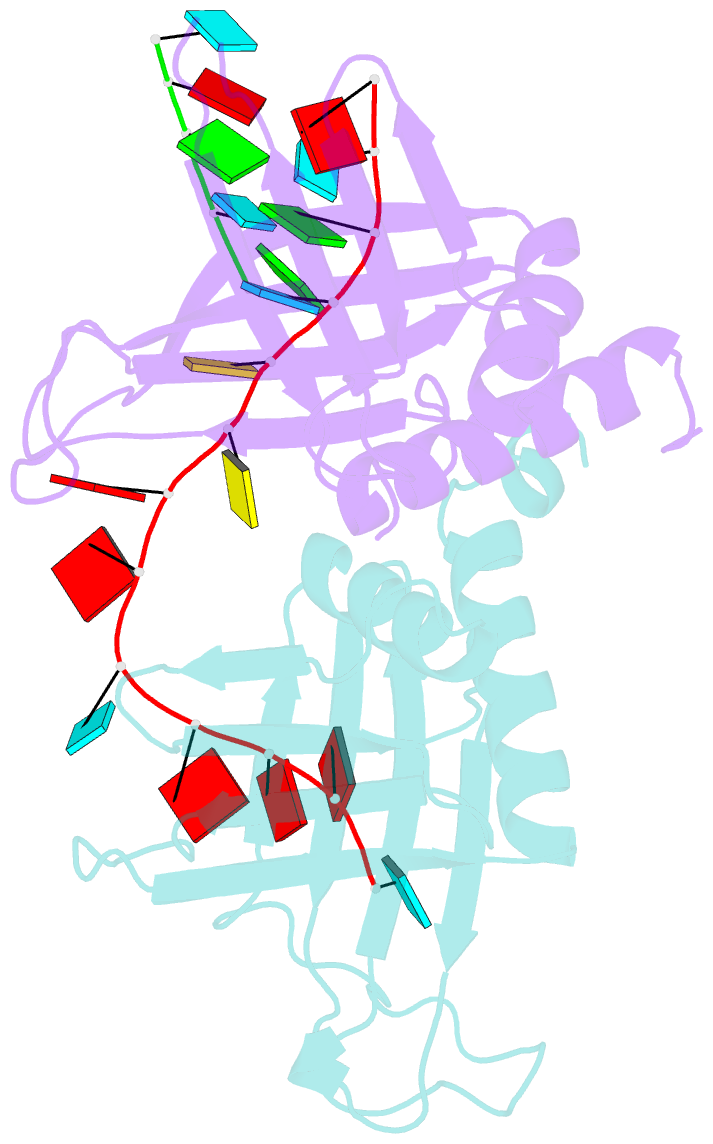
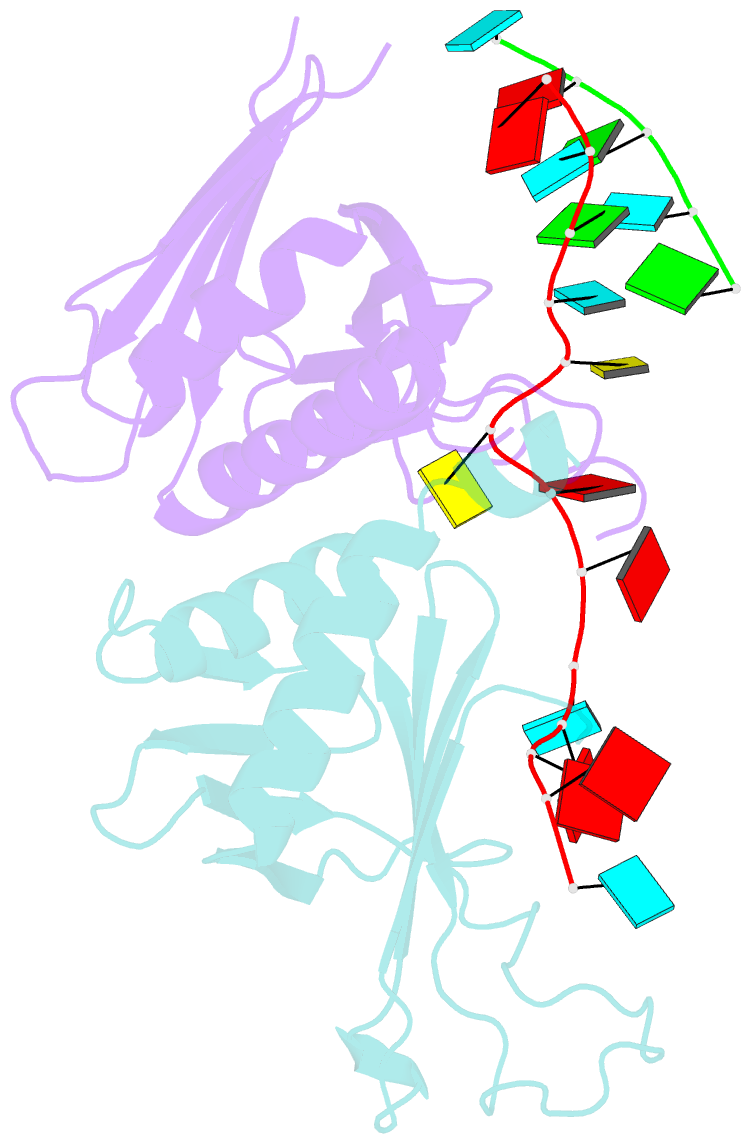
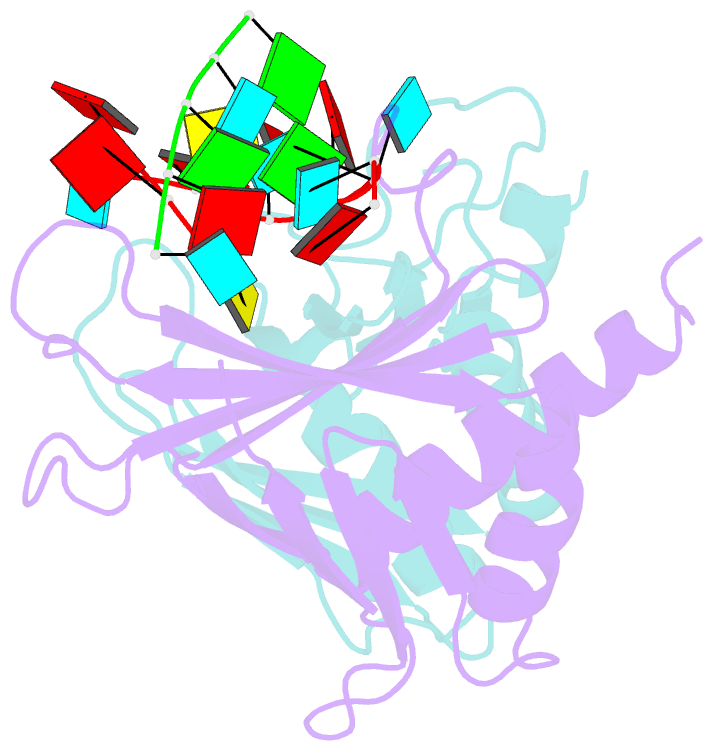
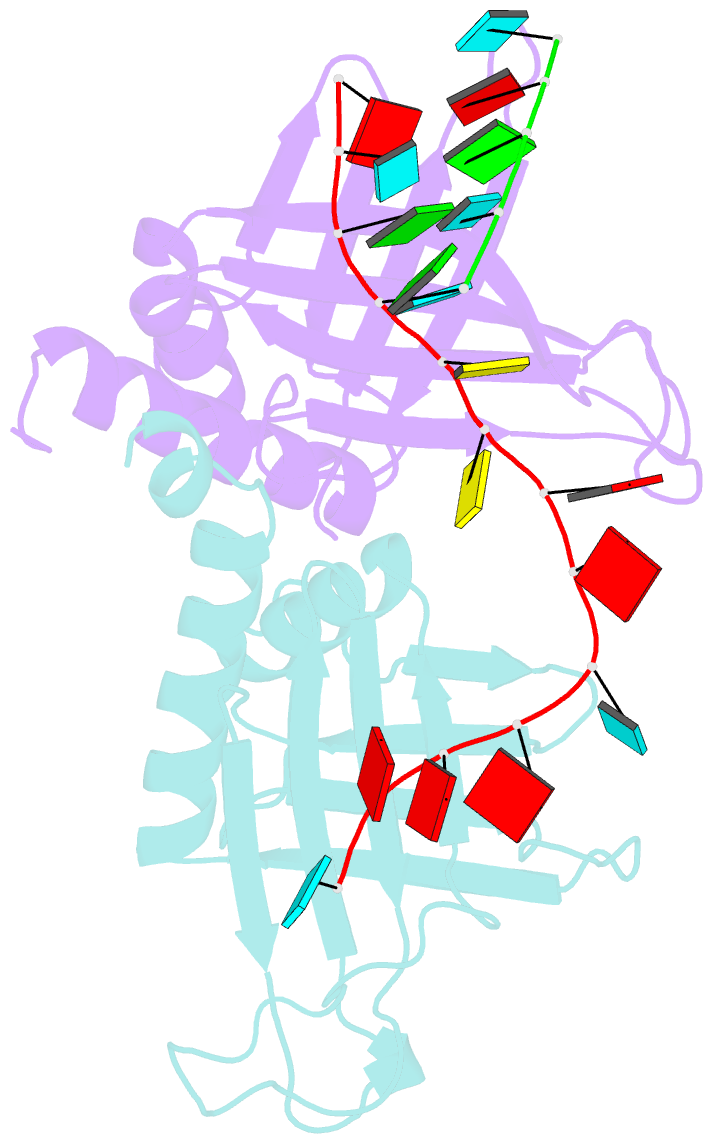
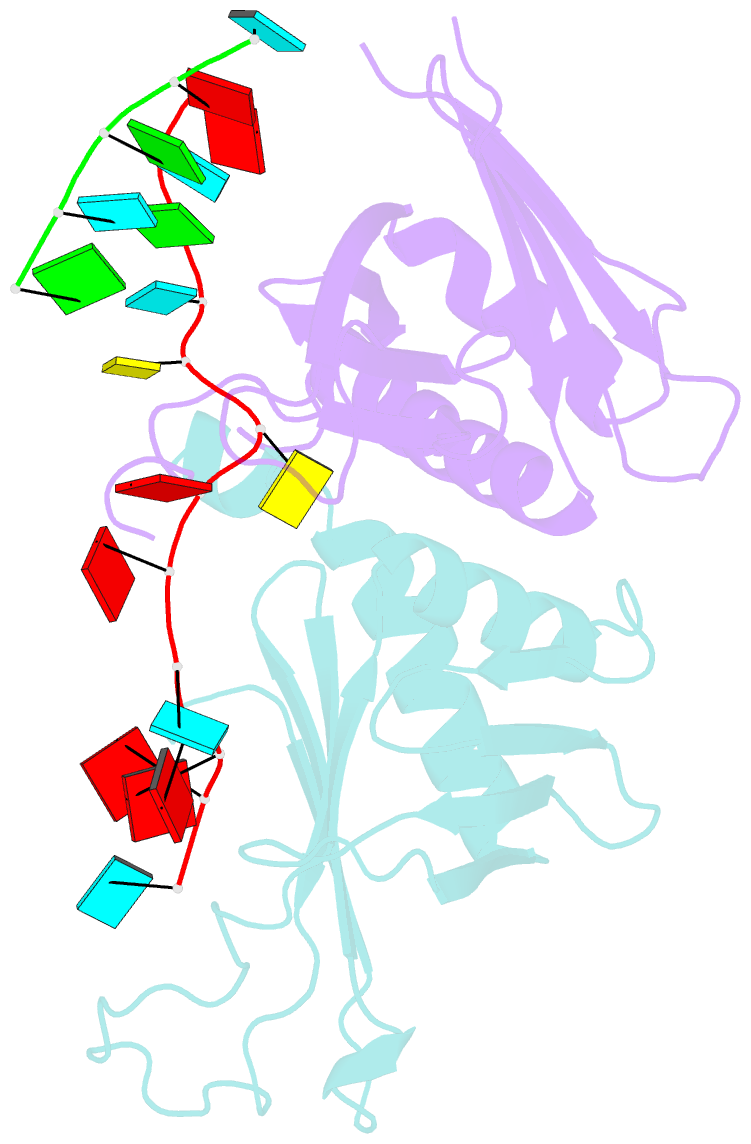
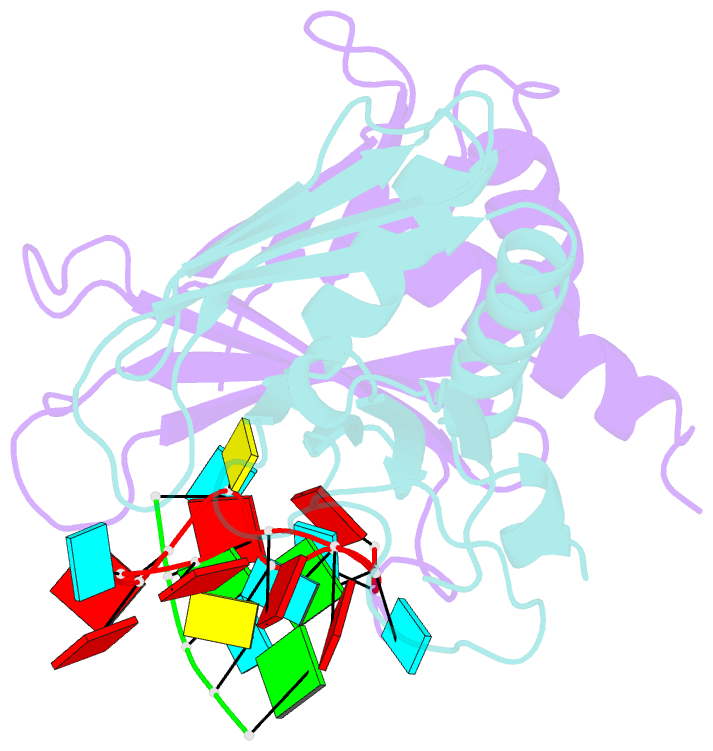
- Summary
- Structure of a guiderna-binding protein complex bound to a grna
- Reference
- Schumacher MA, Karamooz E, Zikova A, Trantirek L, Lukes J (2006): "Crystal Structures of T. brucei MRP1/MRP2 Guide-RNA Binding Complex Reveal RNA Matchmaking Mechanism." Cell(Cambridge,Mass.), 126, 701-711. doi: 10.1016/j.cell.2006.06.047.
- Abstract
- The mitochondrial RNA binding proteins MRP1 and MRP2 form a heteromeric complex that functions in kinetoplastid RNA editing. In this process, MRP1/MRP2 serves as a matchmaker by binding to guide RNAs and facilitating their hybridization with cognate preedited mRNAs. To understand the mechanism by which this complex performs RNA matchmaking, we determined structures of Trypanosoma brucei apoMRP1/MRP2 and an MRP1/MRP2-gRNA complex. The structures show that MRP1/MRP2 is a heterotetramer and, despite little sequence homology, each MRP subunit exhibits the same "Whirly" transcription-factor fold. The gRNA molecule binds to the highly basic beta sheet surface of the MRP complex via nonspecific, electrostatic contacts. Strikingly, while the gRNA stem/loop II base is anchored to the basic surface, stem/loop I (the anchor sequence) is unfolded and its bases exposed to solvent. Thus, MRP1/MRP2 acts as an RNA matchmaker by stabilizing the RNA molecule in an unfolded conformation suitable for RNA-RNA hybridization.