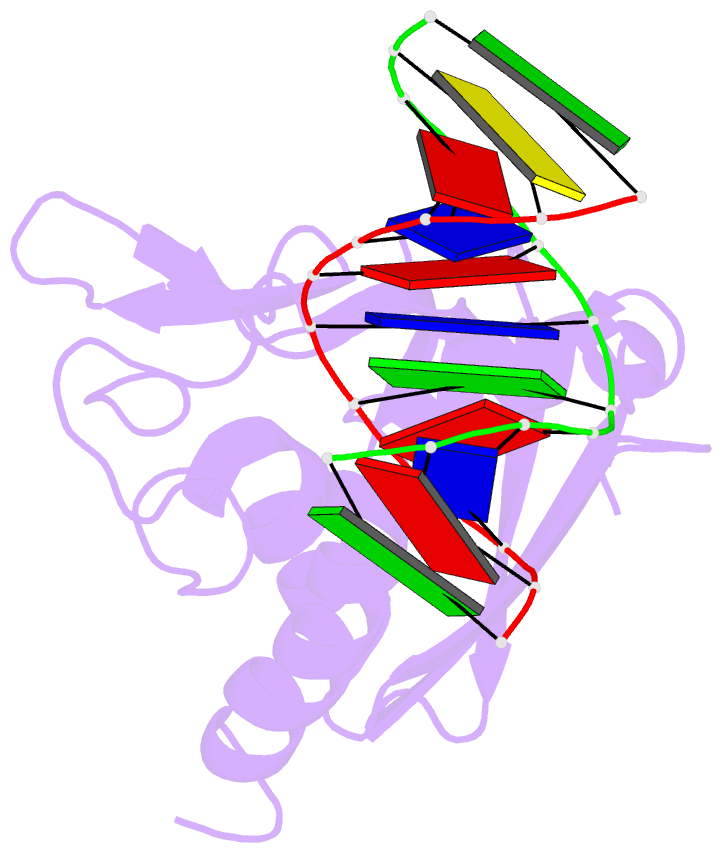
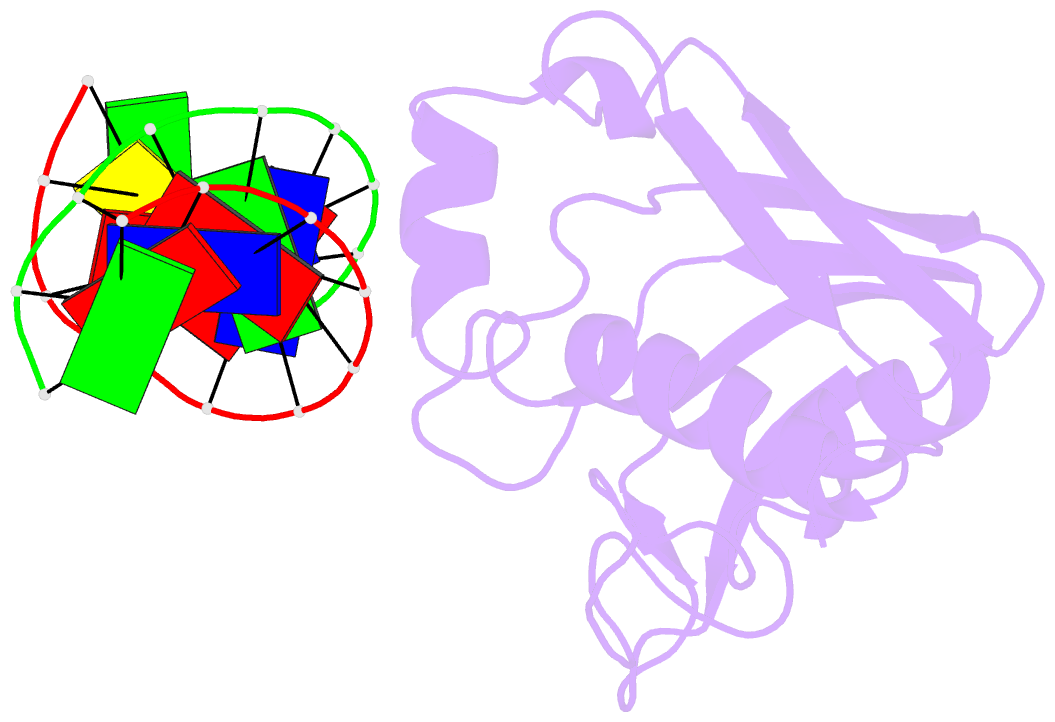
Summary information and primary citation
- PDB-id
- 2gkd; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- toxin-DNA
- Method
- NMR
- Summary
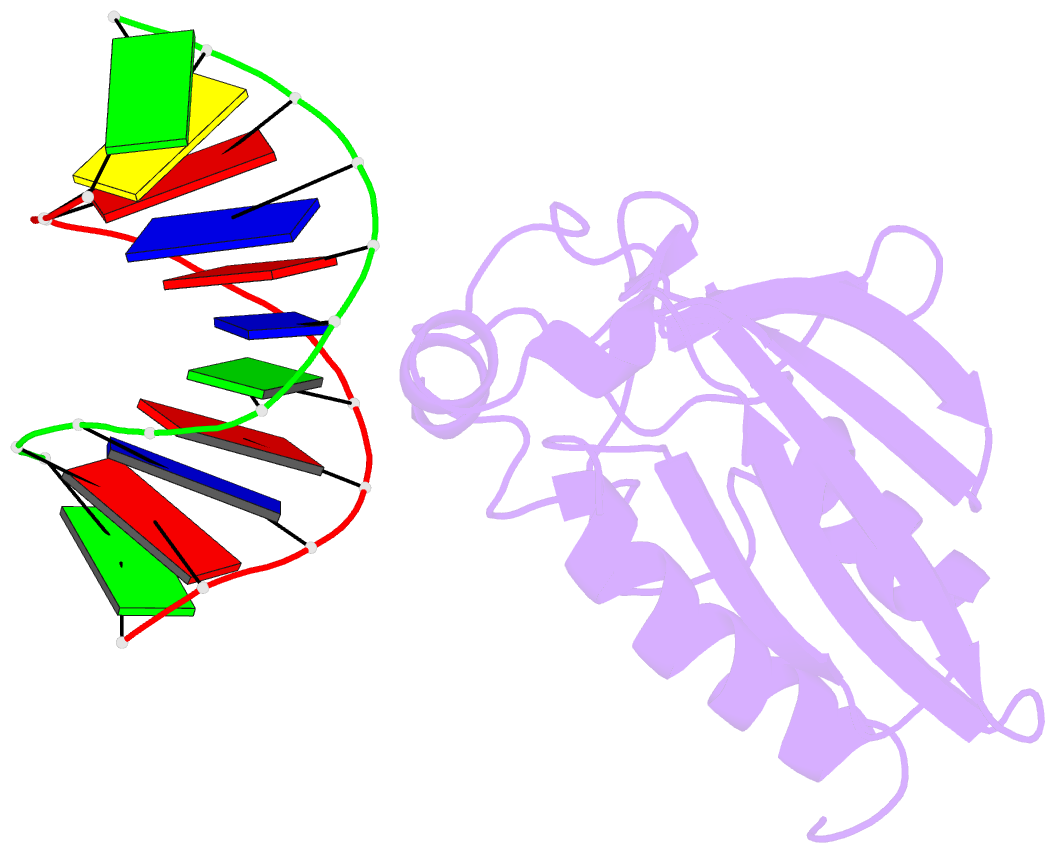
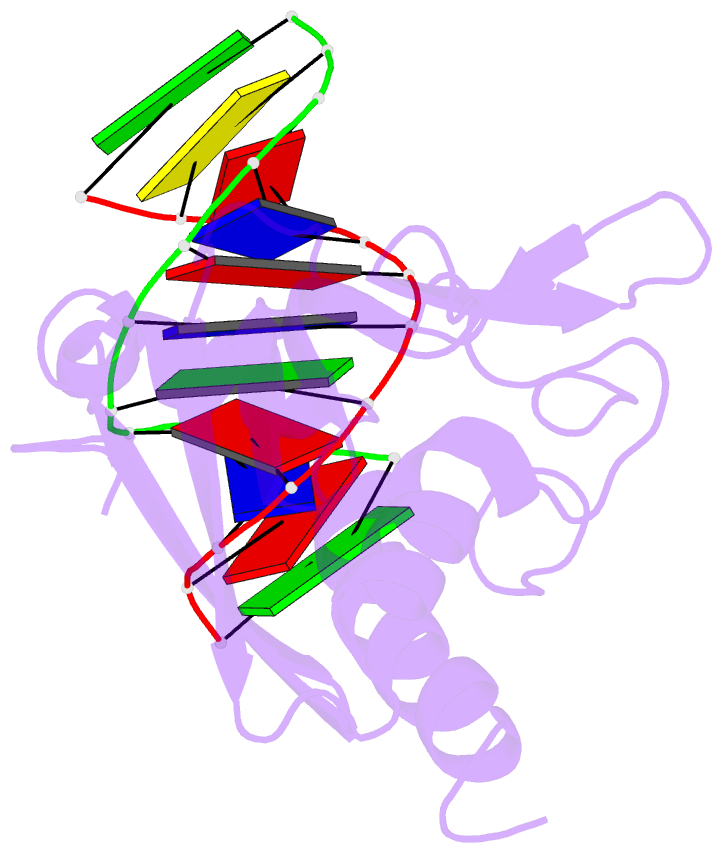
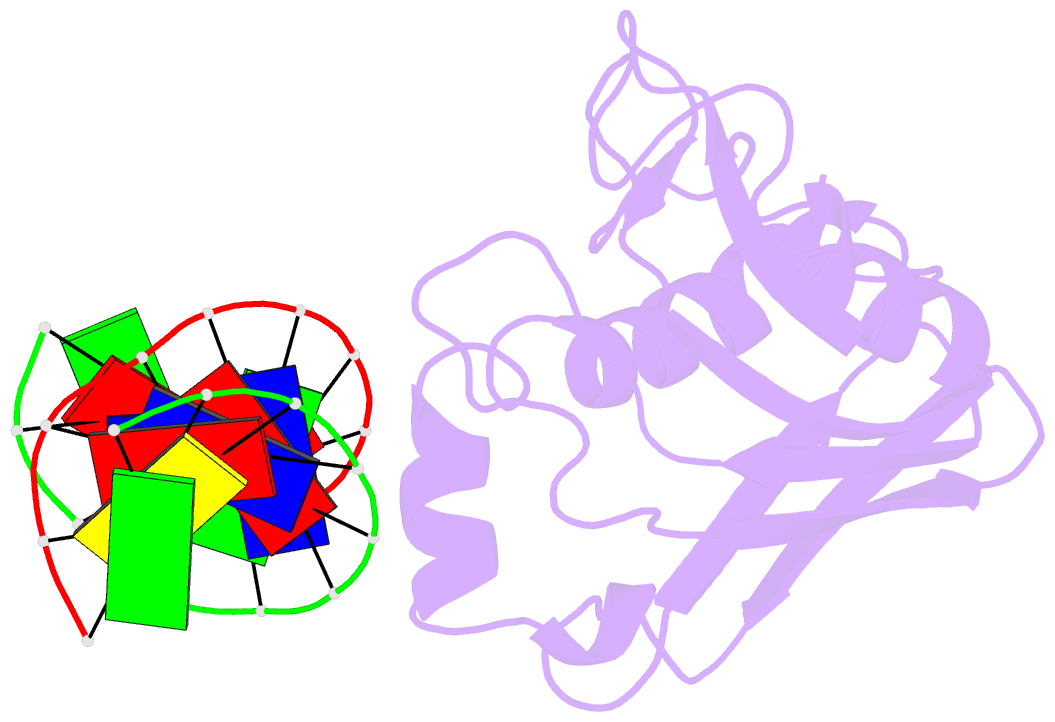
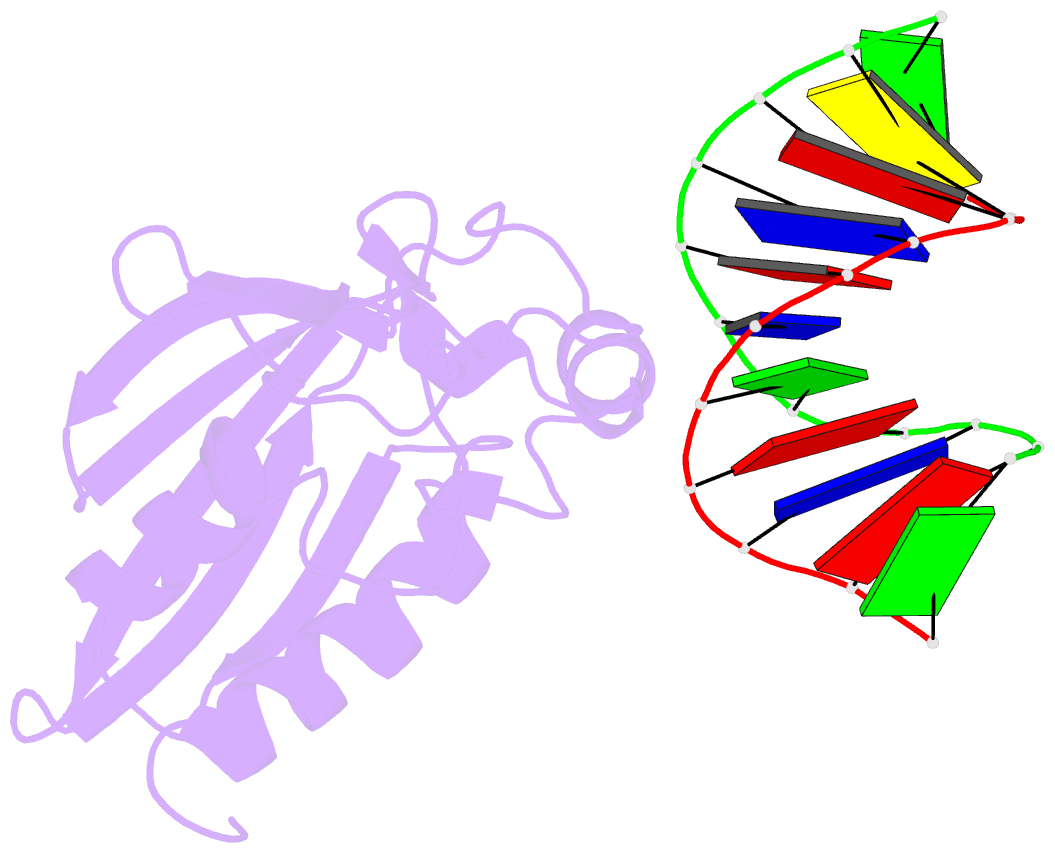
- Structural insight into self-sacrifice mechanism of enediyne resistance
- Reference
- Singh S, Hager MH, Zhang C, Griffith BR, Lee MS, Hallenga K, Markley JL, Thorson JS (2006): "Structural insight into the self-sacrifice mechanism of enediyne resistance." Acs Chem.Biol., 1, 451-460. doi: 10.1021/cb6002898.
- Abstract
- The recent discovery of the first "self-sacrifice" mechanism for bacterial resistance to the enediyne antitumor antibiotics, where enediyne-induced proteolysis of the resistance protein CalC inactivates both the highly reactive metabolite and the resistance protein, revealed yet another ingenious bacterial mechanism for controlling reactive metabolites. As reported herein, the first 3D structures of CalC and CalC in complex with calicheamicin (CLM) divulge CalC to be a member of the steroidogenic acute regulatory protein (StAR)-related transfer (START) domain superfamily. In contrast to previous studies of proteins known to bind DNA-damaging natural products ( e.g ., bleomycins, mitomycins, and nine-membered chromoprotein enediynes), this is the first demonstrated involvement of a START domain fold. Consistent with the CalC self-sacrifice mechanism, CLM in complex with CalC is positioned for direct hydrogen abstraction from Gly113 to initiate the oxidative proteolysis-based resistance mechanism. These structural studies also illuminate, for the first time, a small DNA-binding region within CalC that may serve to localize CalC to the enediyne target (DNA). Given the role of START domains in nuclear/cytosolic transport and translocation, this structural study also may implicate START domains as post-endocytotic intracellular chaperones for enediyne-based therapeutics such as MyloTarg.