Summary information and primary citation
- PDB-id
- 2glo; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- NMR
- Summary
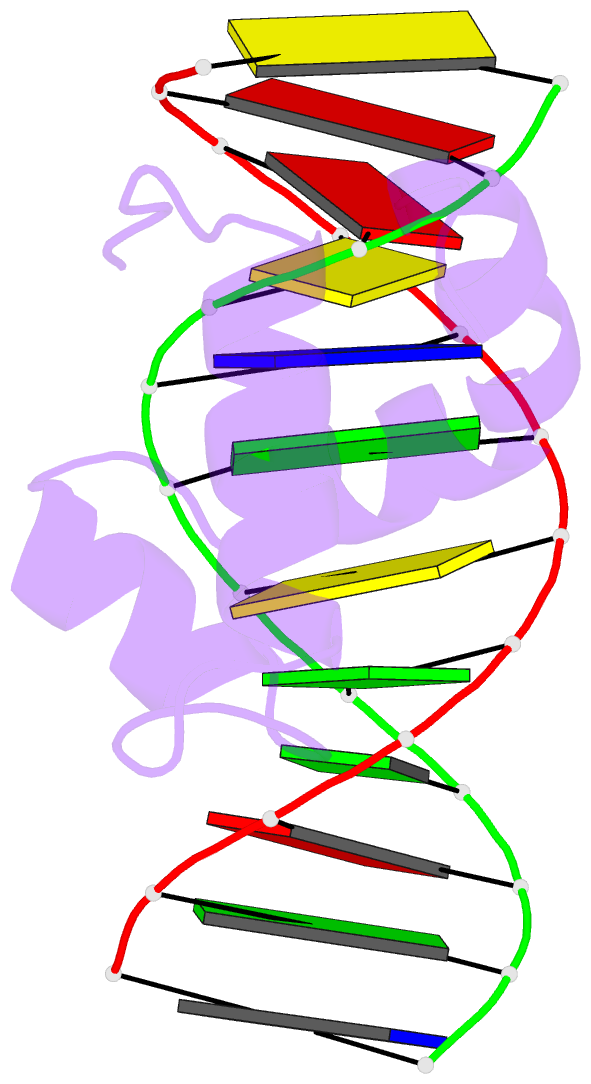
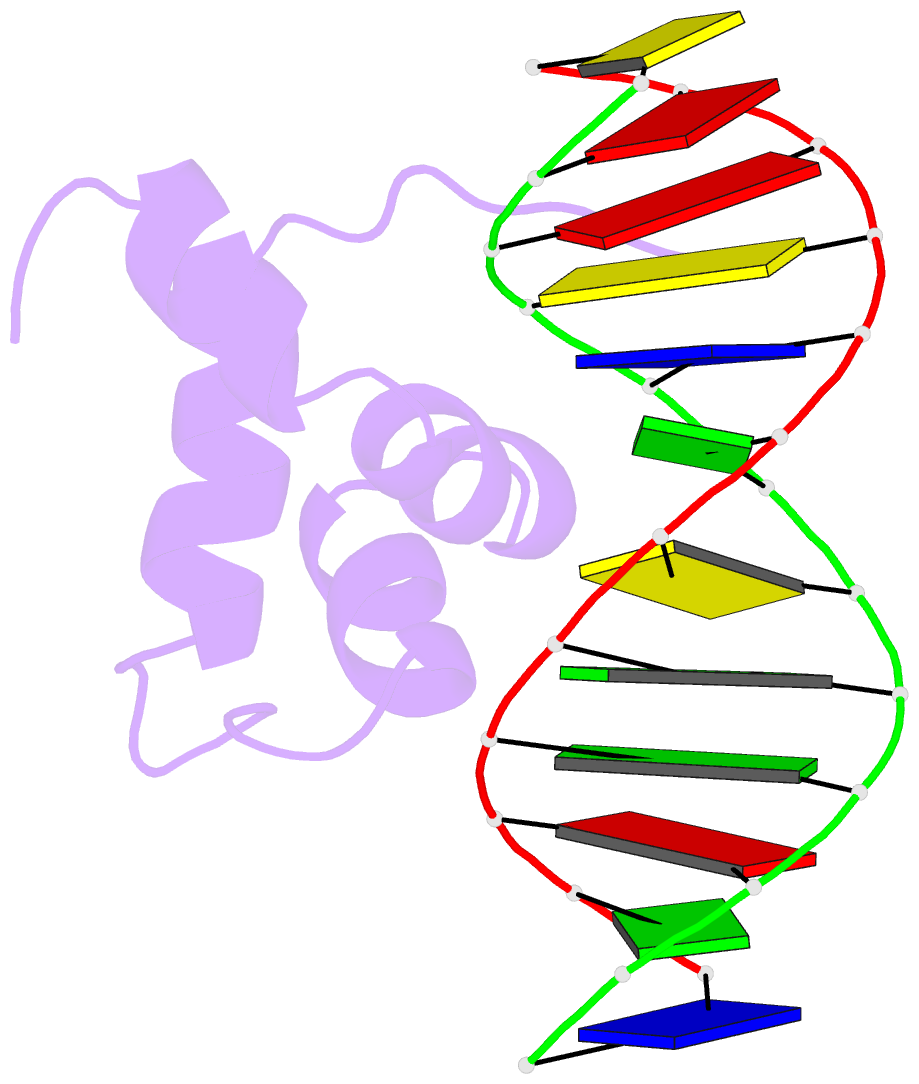
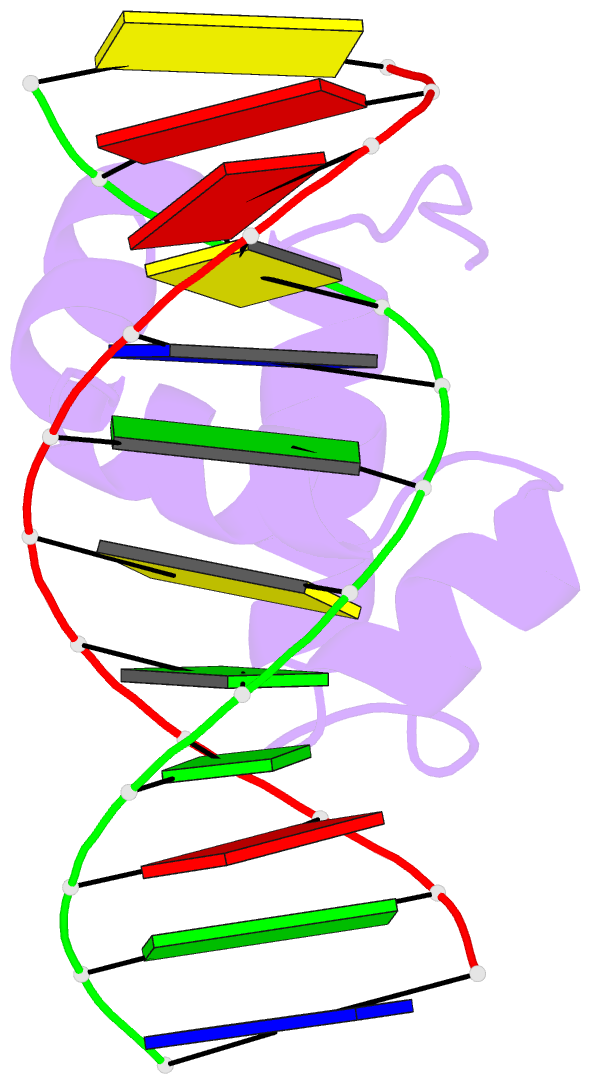
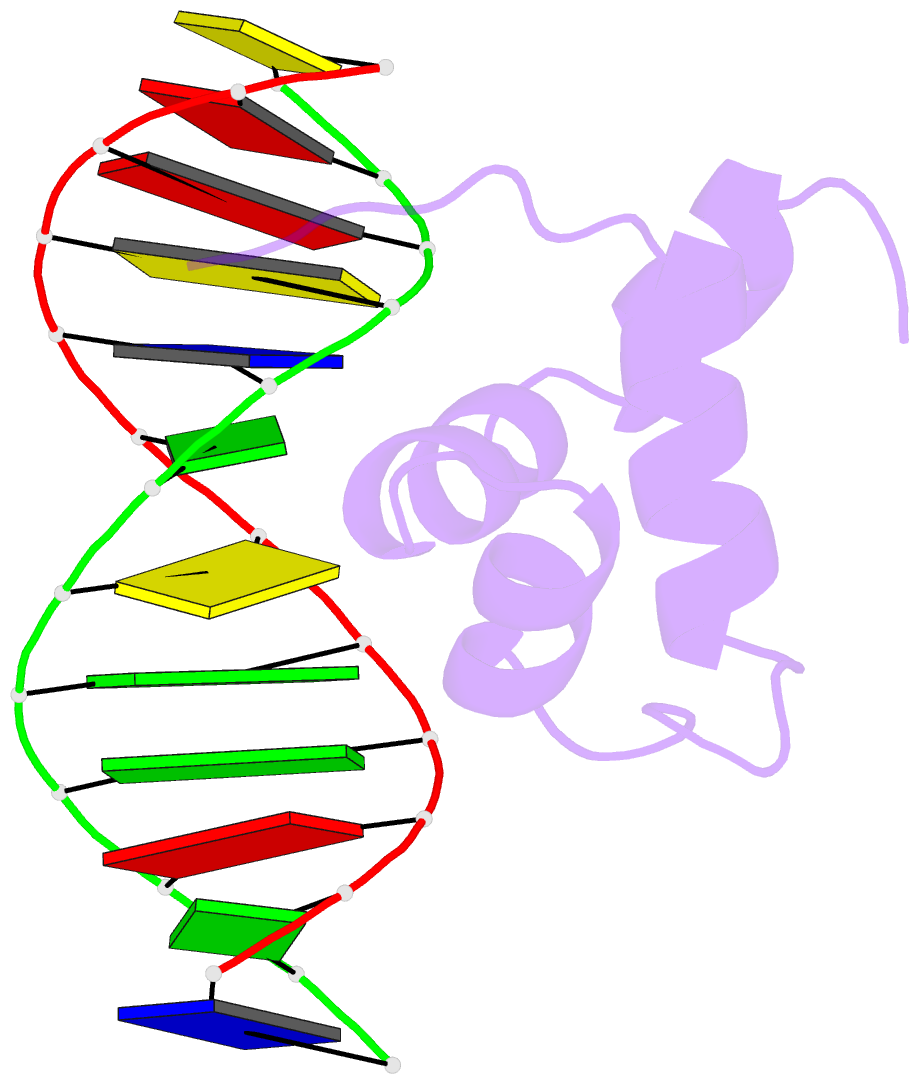
- Solution structure of the brinker DNA binding domain in complex with the omb enhancer
- Reference
- Cordier F, Hartmann B, Rogowski M, Affolter M, Grzesiek S (2006): "DNA recognition by the brinker repressor - an extreme case of coupling between binding and folding." J.Mol.Biol., 361, 659-672. doi: 10.1016/j.jmb.2006.06.045.
- Abstract
- The Brinker (Brk) nuclear repressor is a major element of the Drosophila Decapentaplegic morphogen signaling pathway. Its N-terminal part has weak homology to the Antennapedia homeodomain and binds to GC-rich DNA sequences. We have investigated the conformation and dynamics of the N-terminal 101 amino acid residues of Brk in the absence and in the presence of cognate DNA by solution NMR spectroscopy. In the absence of DNA, Brk is unfolded and highly flexible throughout the entire backbone. Addition of cognate DNA induces the formation of a well-folded structure for residues R46 to R95. This structure consists of four helices forming a helix-turn-helix motif that differs from homeodomains, but has similarities to the Tc3 transposase, the Pax-6 Paired domain, and the human centromere-binding protein. The GC-rich DNA recognition can be explained by specific major groove hydrogen bonds from the N-terminal end of helix alpha3. The transition from a highly flexible, completely unfolded conformation in the absence of DNA to a well-formed structure in the complex presents a very extreme case of the "coupling of binding and folding" phenomenon.