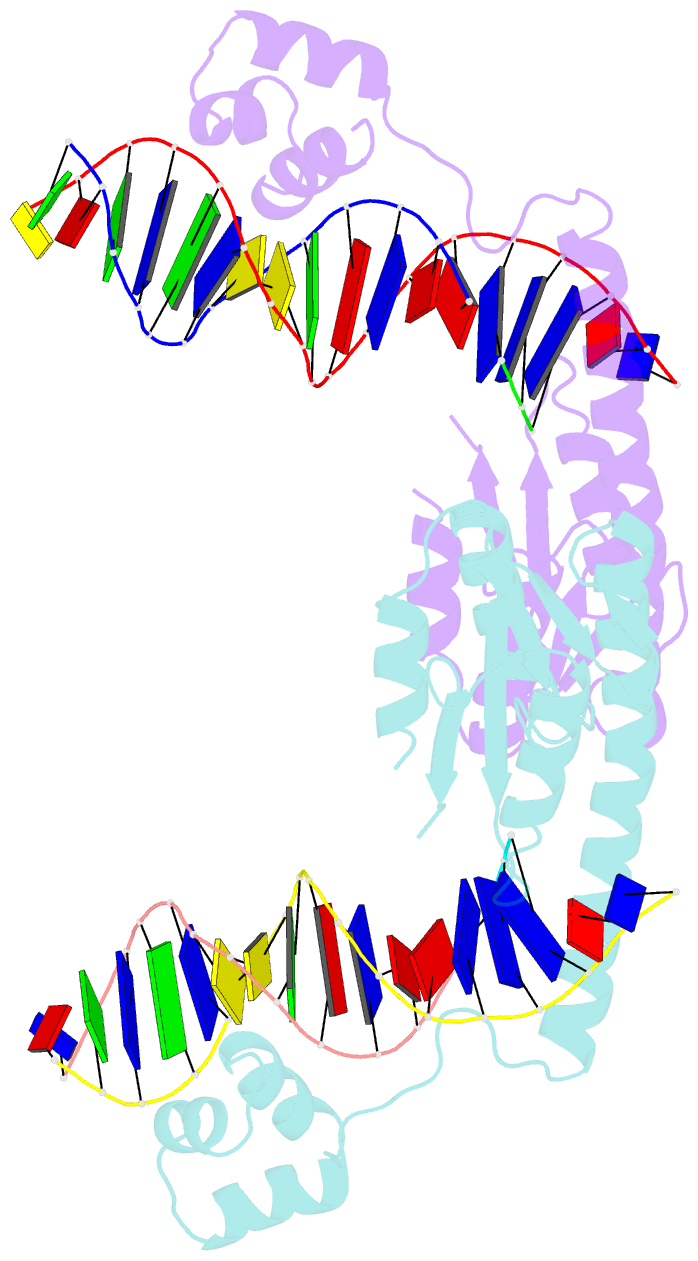
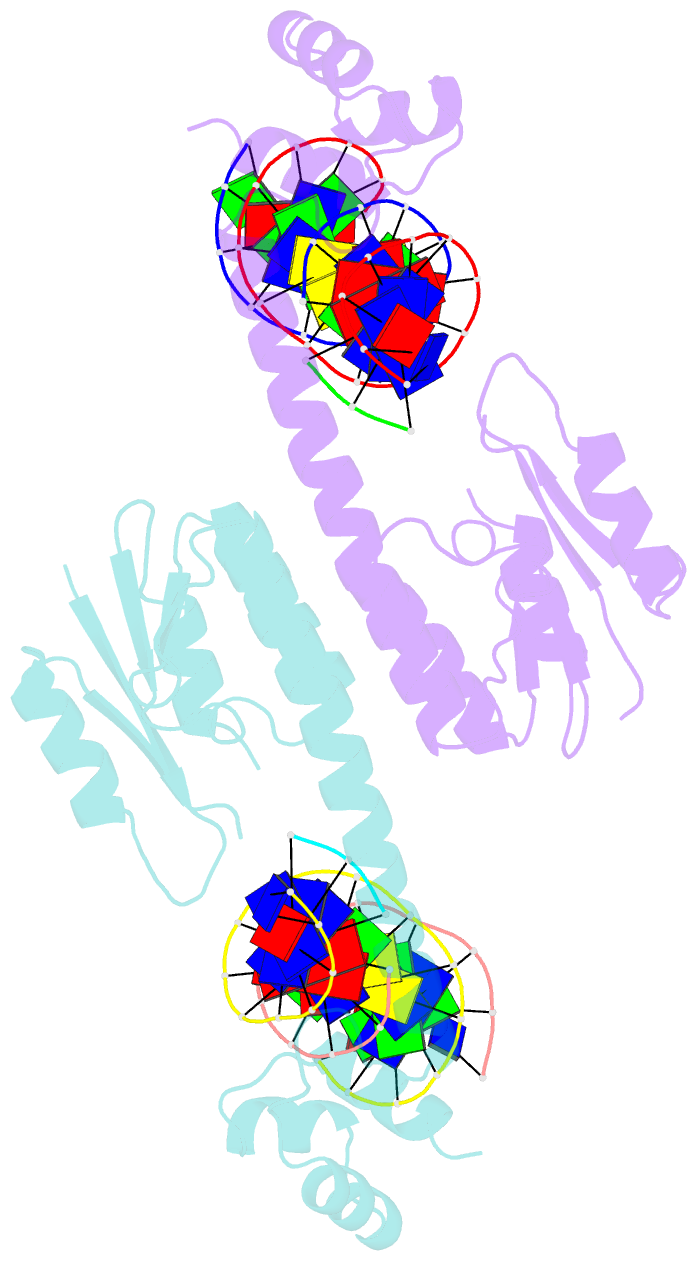
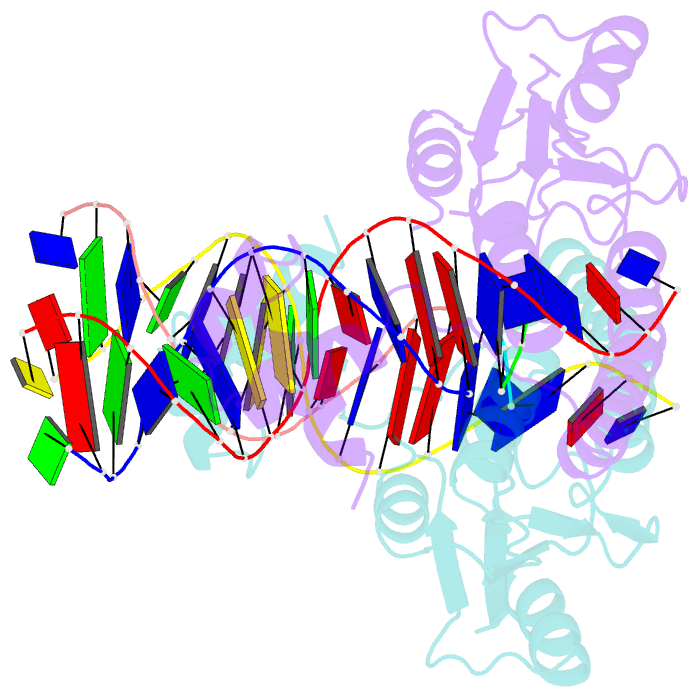
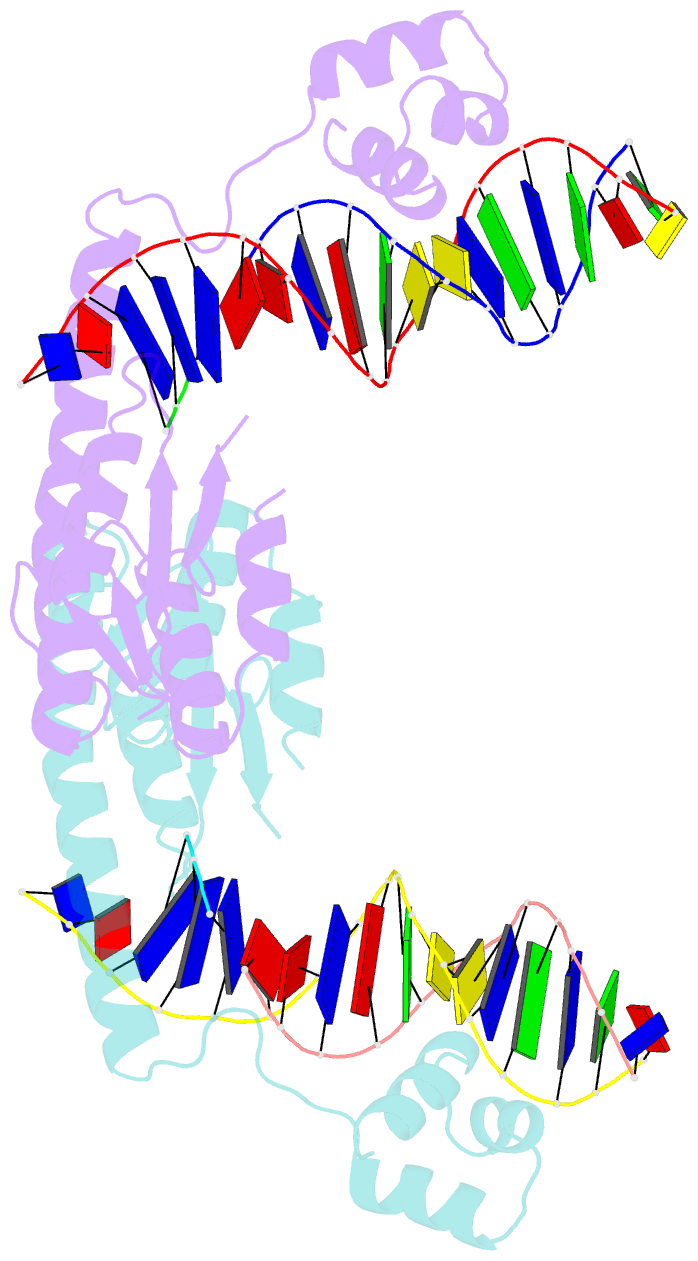
Summary information and primary citation
- PDB-id
- 2gm4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- recombination, DNA
- Method
- X-ray (3.5 Å)
- Summary
- An activated, tetrameric gamma-delta resolvase: hin chimaera bound to cleaved DNA
- Reference
- Kamtekar S, Ho RS, Cocco MJ, Li W, Wenwieser SV, Boocock MR, Grindley ND, Steitz TA (2006): "Implications of structures of synaptic tetramers of gamma delta resolvase for the mechanism of recombination." Proc.Natl.Acad.Sci.Usa, 103, 10642-10647. doi: 10.1073/pnas.0604062103.
- Abstract
- The structures of two mutants of the site-specific recombinase, gammadelta resolvase, that form activated tetramers have been determined. One, at 3.5-A resolution, forms a synaptic intermediate of resolvase that is covalently linked to two cleaved DNAs, whereas the other is of an unliganded structure determined at 2.1-A resolution. Comparisons of the four known tetrameric resolvase structures show that the subunits interact through the formation of a common core of four helices. The N-terminal halves of these helices superimpose well on each other, whereas the orientations of their C termini are more variable. The catalytic domains of resolvase in the unliganded structure are arranged asymmetrically, demonstrating that their positions can move substantially while preserving the four-helix core that forms the tetramer. These results suggest that the precleavage synaptic tetramer of gammadelta resolvase, whose structure is not known, may be formed by a similar four-helix core, but differ in the relative orientations of its catalytic and DNA-binding domains.