Summary information and primary citation
- PDB-id
- 2hw8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- structural protein-RNA
- Method
- X-ray (2.1 Å)
- Summary
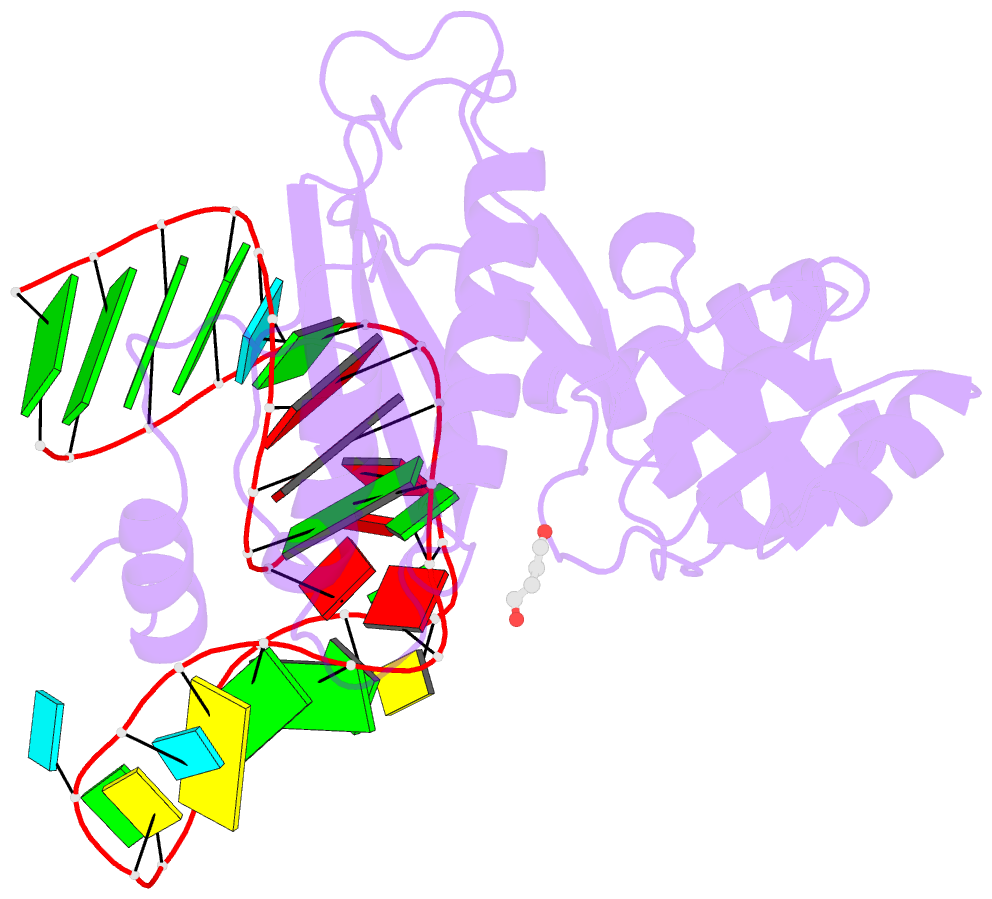
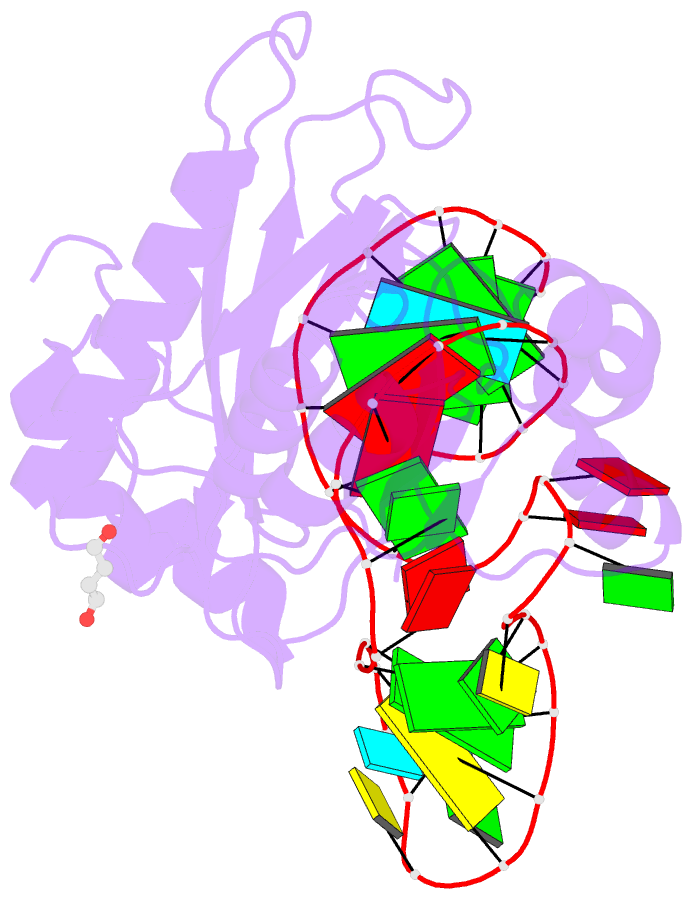
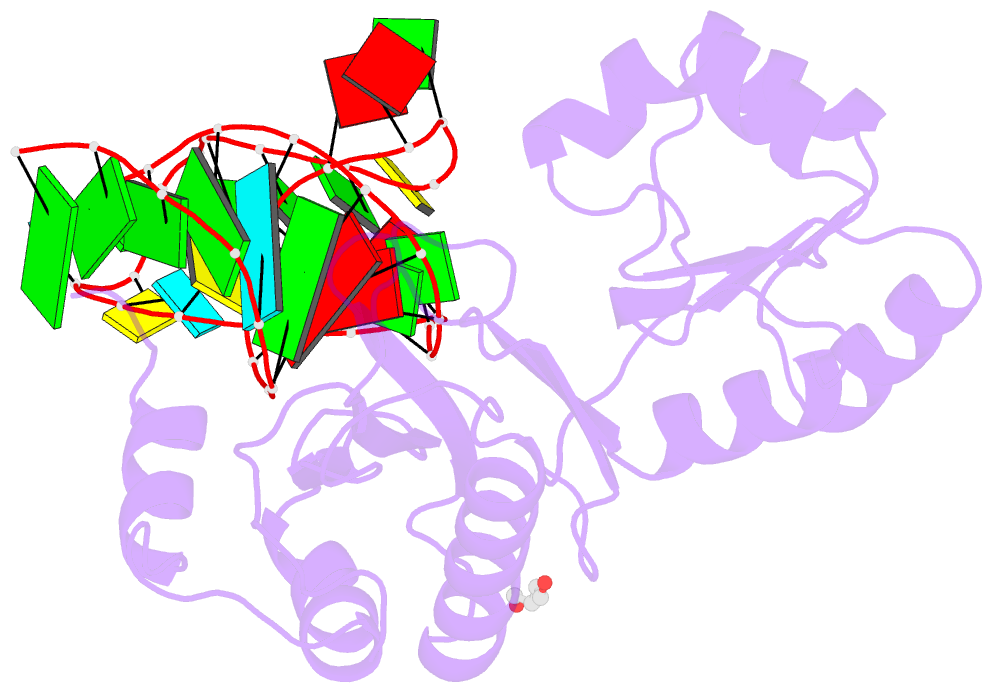
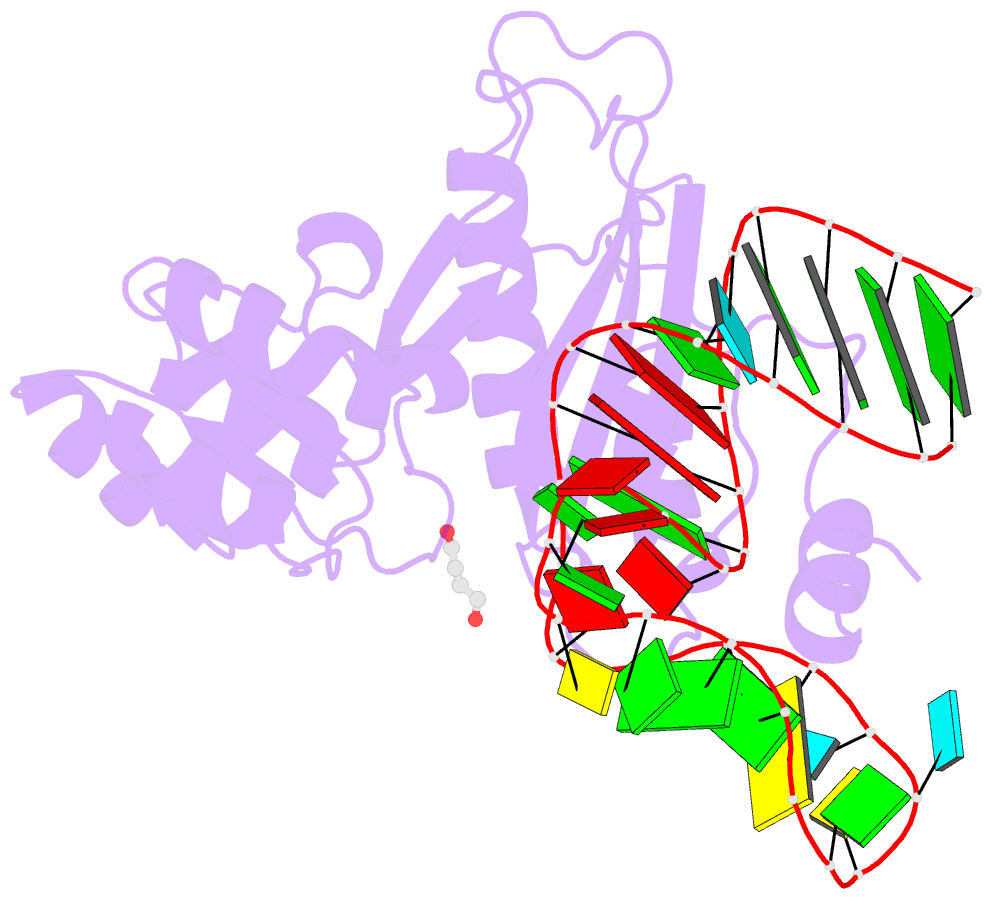
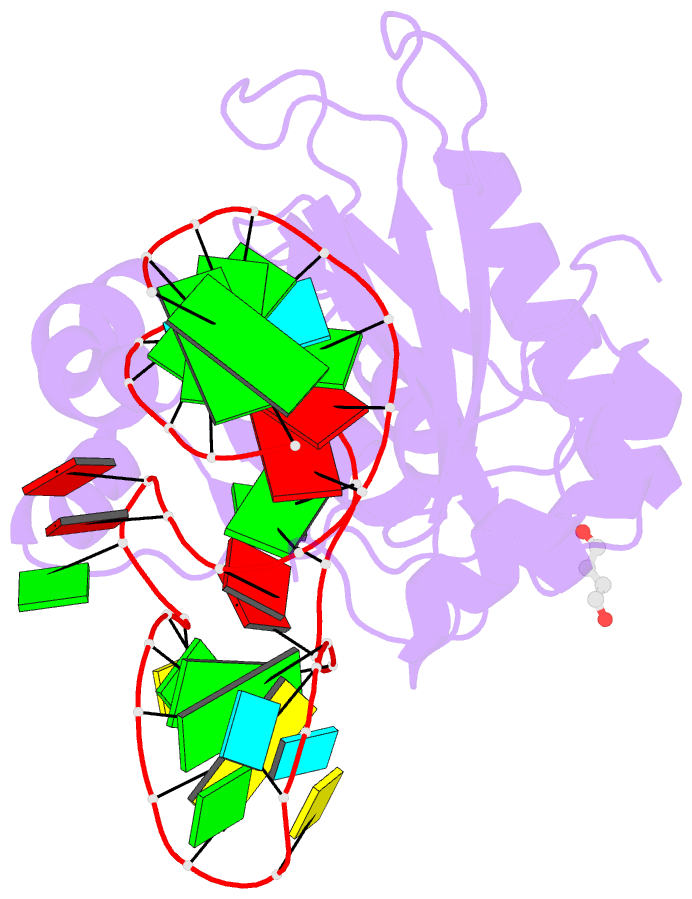
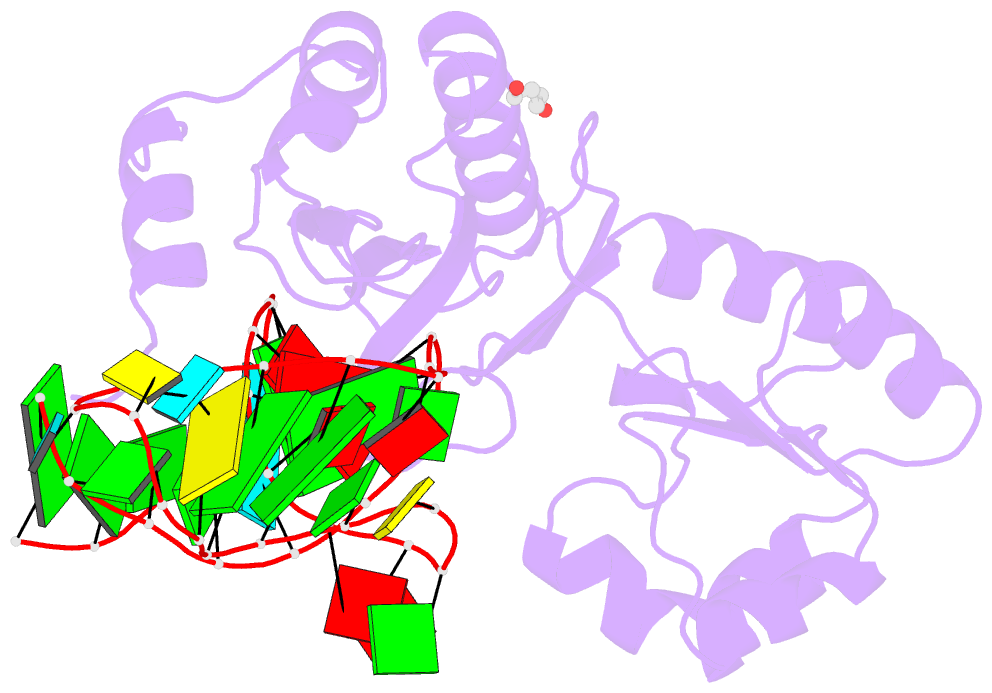
- Structure of ribosomal protein l1-mrna complex at 2.1 resolution.
- Reference
- Tishchenko S, Nikonova E, Nikulin A, Nevskaya N, Volchkov S, Piendl W, Garber M, Nikonov S (2006): "Structure of the ribosomal protein L1-mRNA complex at 2.1 A resolution: common features of crystal packing of L1-RNA complexes." ACTA CRYSTALLOGR.,SECT.D, 62, 1545-1554. doi: 10.1107/S0907444906041655.
- Abstract
- The crystal structure of a hybrid complex between the bacterial ribosomal protein L1 from Thermus thermophilus and a Methanococcus vannielii mRNA fragment containing an L1-binding site was determined at 2.1 A resolution. It was found that all polar atoms involved in conserved protein-RNA hydrogen bonds have high values of density in the electron-density map and that their hydrogen-bonding capacity is fully realised through interactions with protein atoms, water molecules and K(+) ions. Intermolecular contacts were thoroughly analyzed in the present crystals and in crystals of previously determined L1-RNA complexes. It was shown that extension of the RNA helices providing canonical helix stacking between open-open or open-closed ends of RNA fragments is a common feature of these and all known crystals of complexes between ribosomal proteins and RNAs. In addition, the overwhelming majority of complexes between ribosomal proteins and RNA molecules display crystal contacts formed by the central parts of the RNA fragments. These contacts are often very extensive and strong and it is proposed that they are formed in the saturated solution prior to crystal formation.