Summary information and primary citation
- PDB-id
- 2hzv; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- metal binding protein-DNA
- Method
- X-ray (3.1 Å)
- Summary
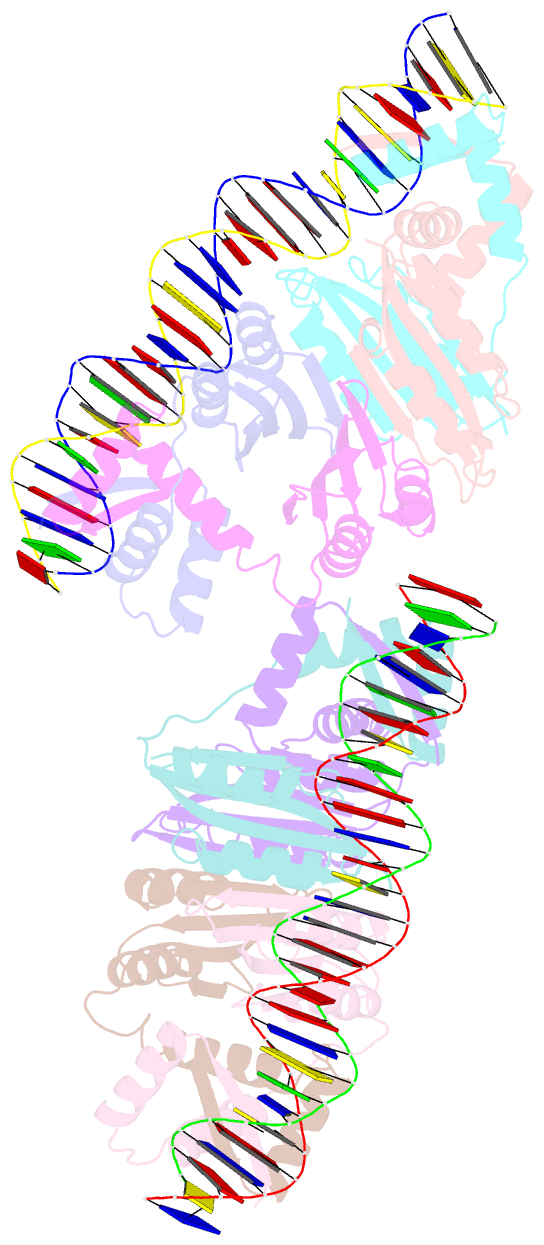
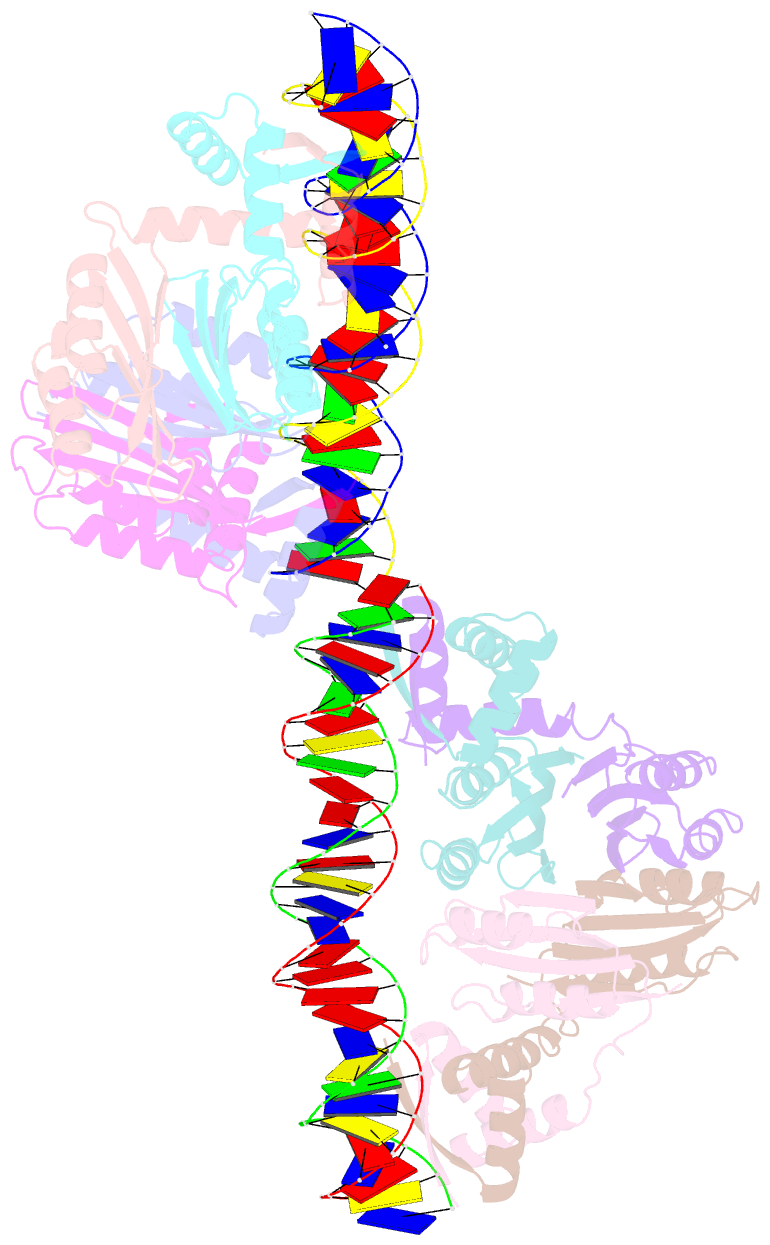
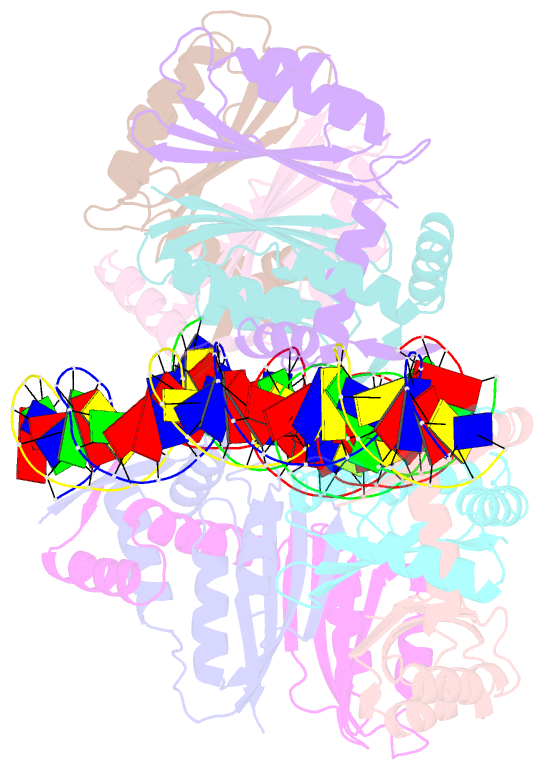
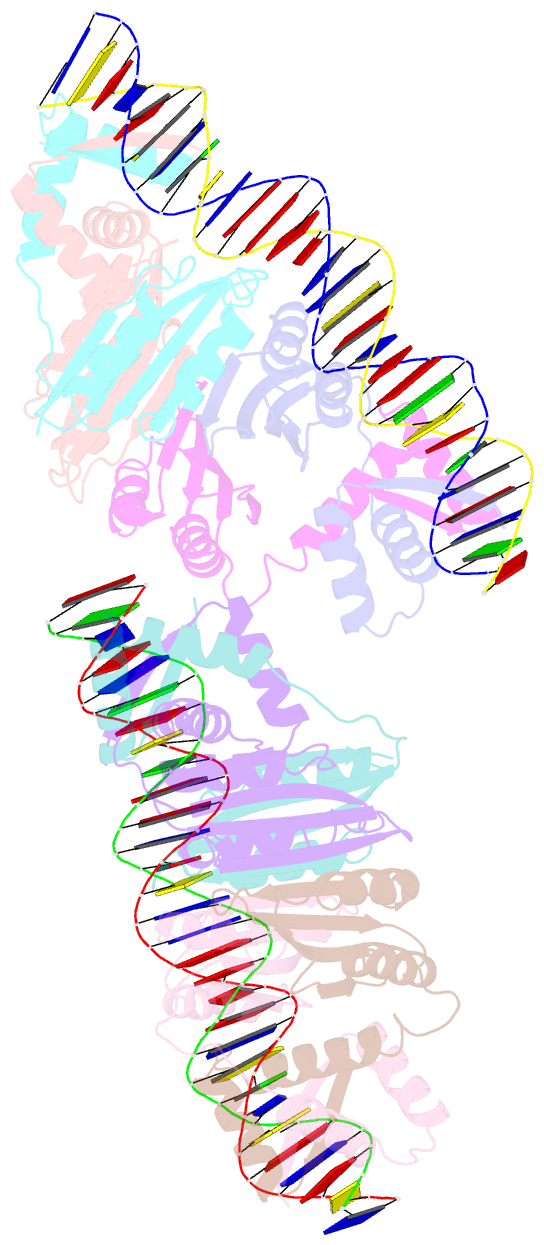
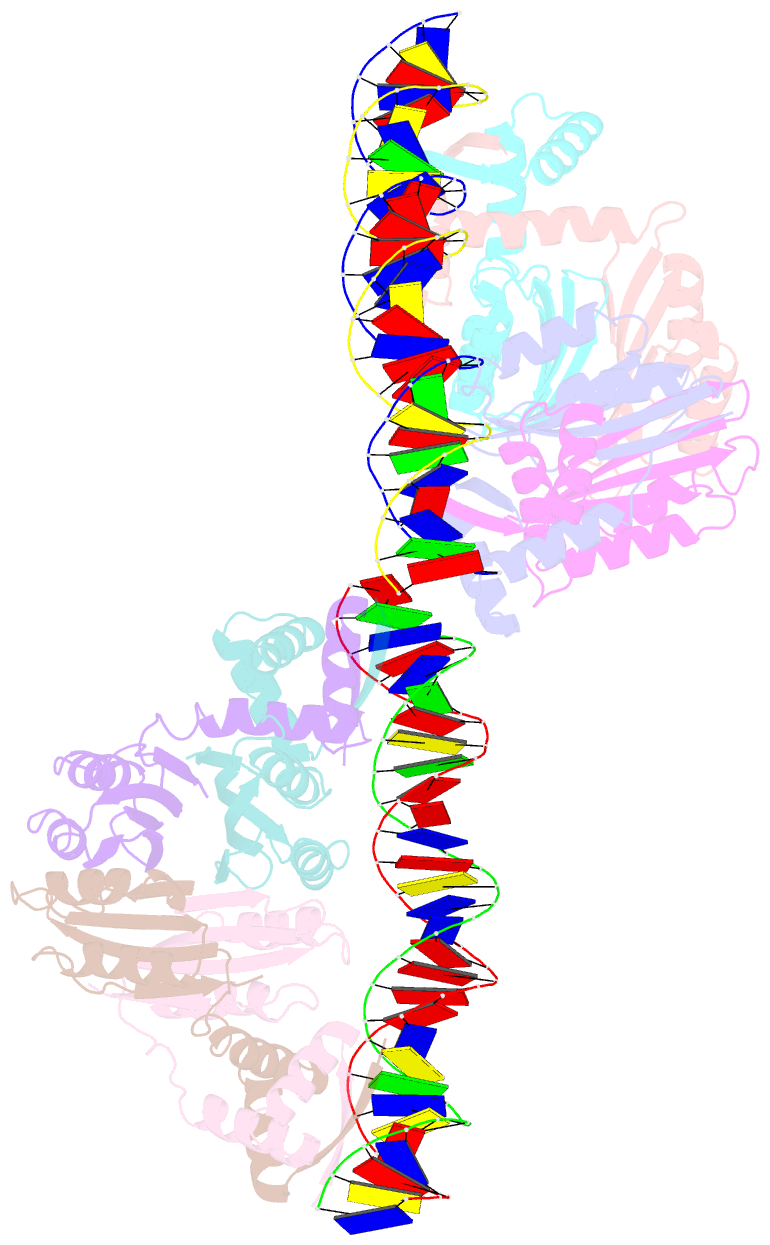
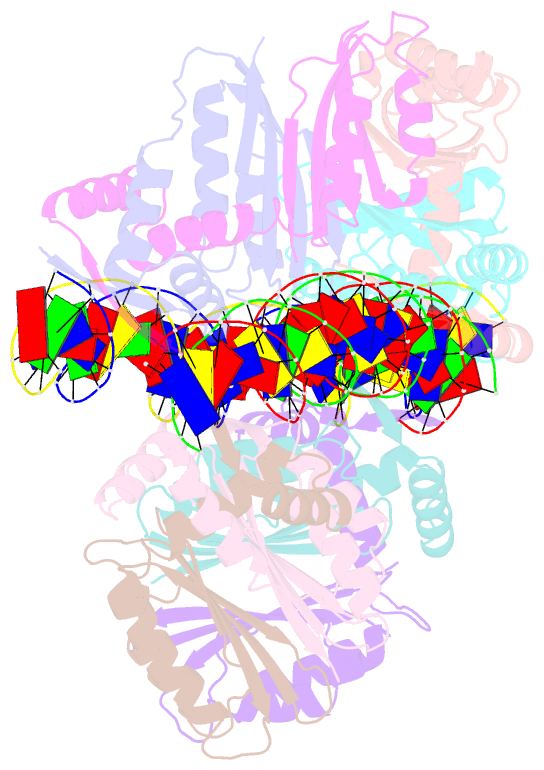
- Nikr-operator DNA complex
- Reference
- Schreiter ER, Wang SC, Zamble DB, Drennan CL (2006): "NikR-operator complex structure and the mechanism of repressor activation by metal ions." Proc.Natl.Acad.Sci.Usa, 103, 13676-13681. doi: 10.1073/pnas.0606247103.
- Abstract
- Metal ion homeostasis is critical to the survival of all cells. Regulation of nickel concentrations in Escherichia coli is mediated by the NikR repressor via nickel-induced transcriptional repression of the nickel ABC-type transporter, NikABCDE. Here, we report two crystal structures of nickel-activated E. coli NikR, the isolated repressor at 2.1 A resolution and in a complex with its operator DNA sequence from the nik promoter at 3.1 A resolution. Along with the previously published structure of apo-NikR, these structures allow us to evaluate functional proposals for how metal ions activate NikR, delineate the drastic conformational changes required for operator recognition, and describe the formation of a second metal-binding site in the presence of DNA. They also provide a rare set of structural views of a ligand-responsive transcription factor in the unbound, ligand-induced, and DNA-bound states, establishing a model system for the study of ligand-mediated effects on transcription factor function.