Summary information and primary citation
- PDB-id
- 2i5w; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase, lyase-DNA
- Method
- X-ray (2.6 Å)
- Summary
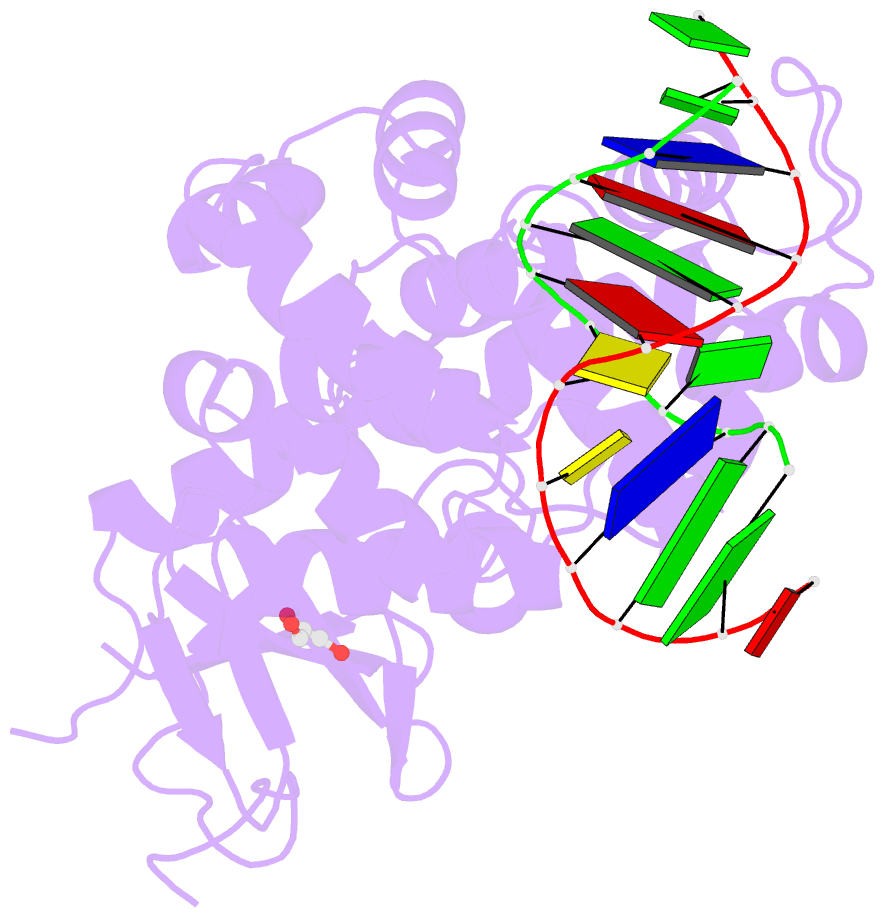
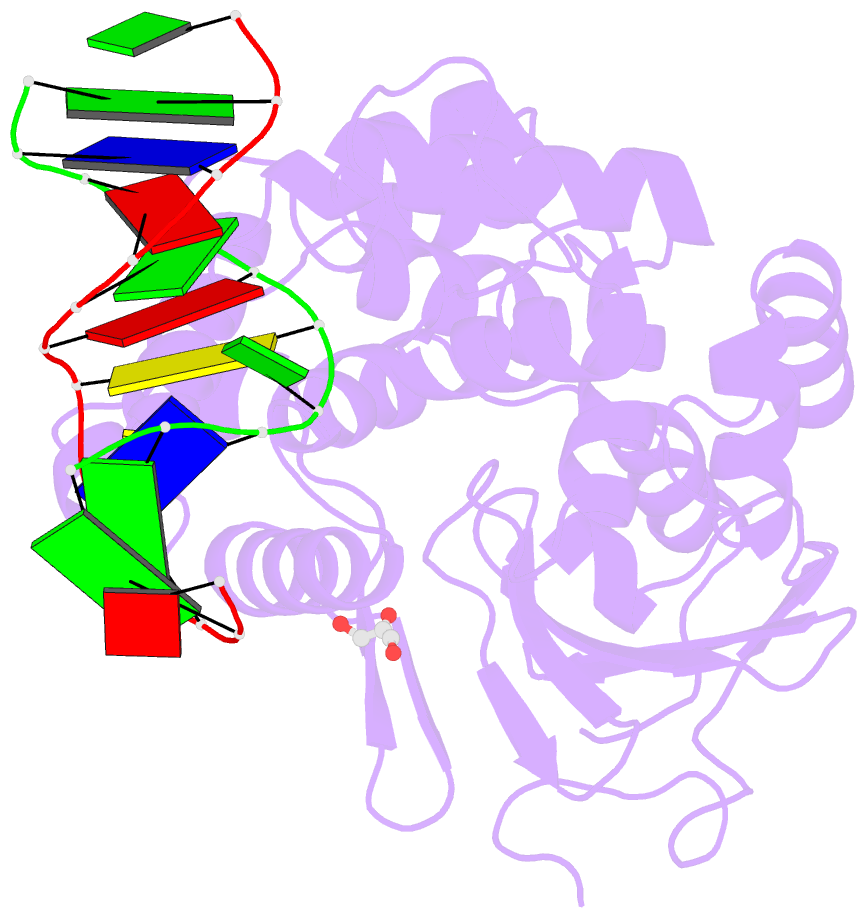
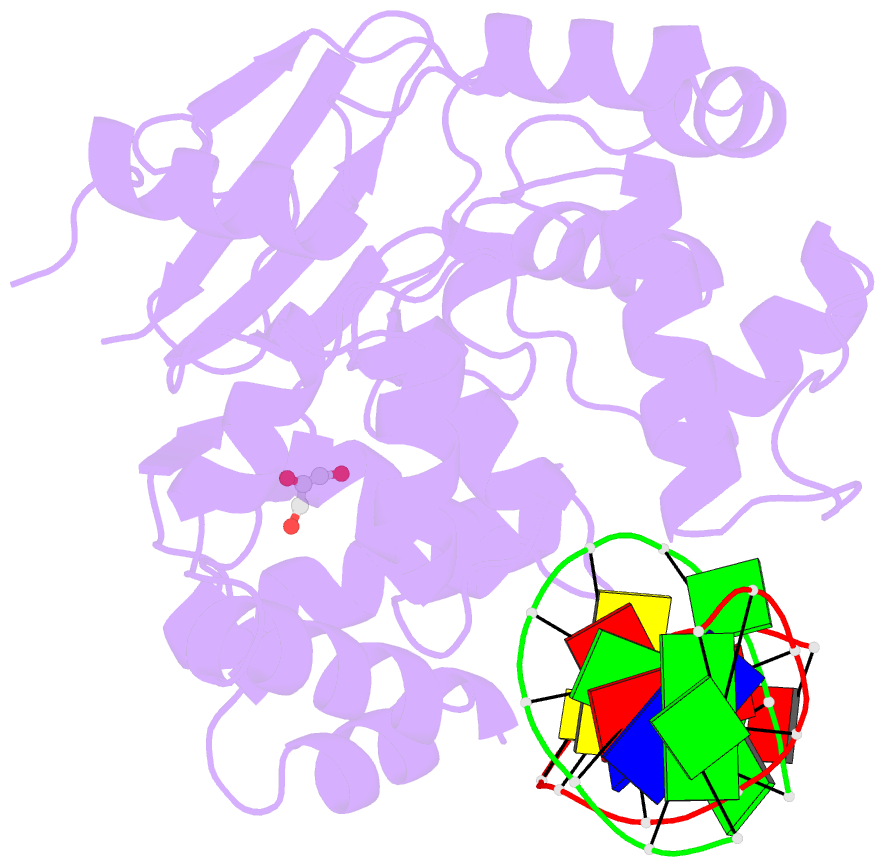
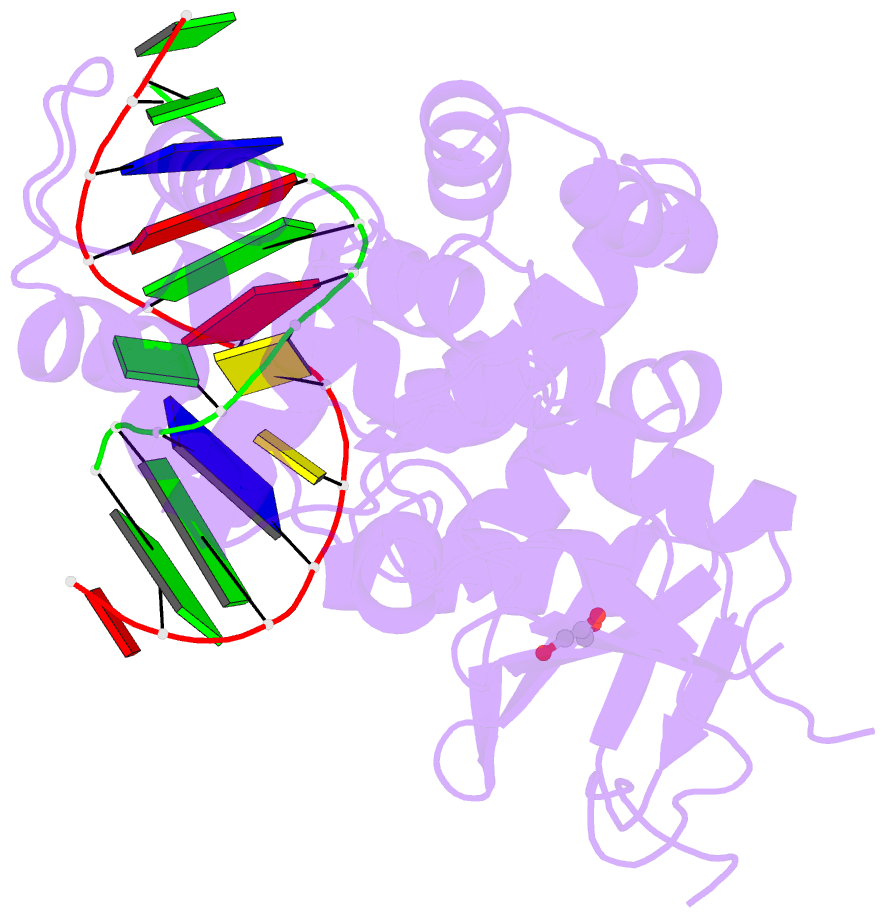
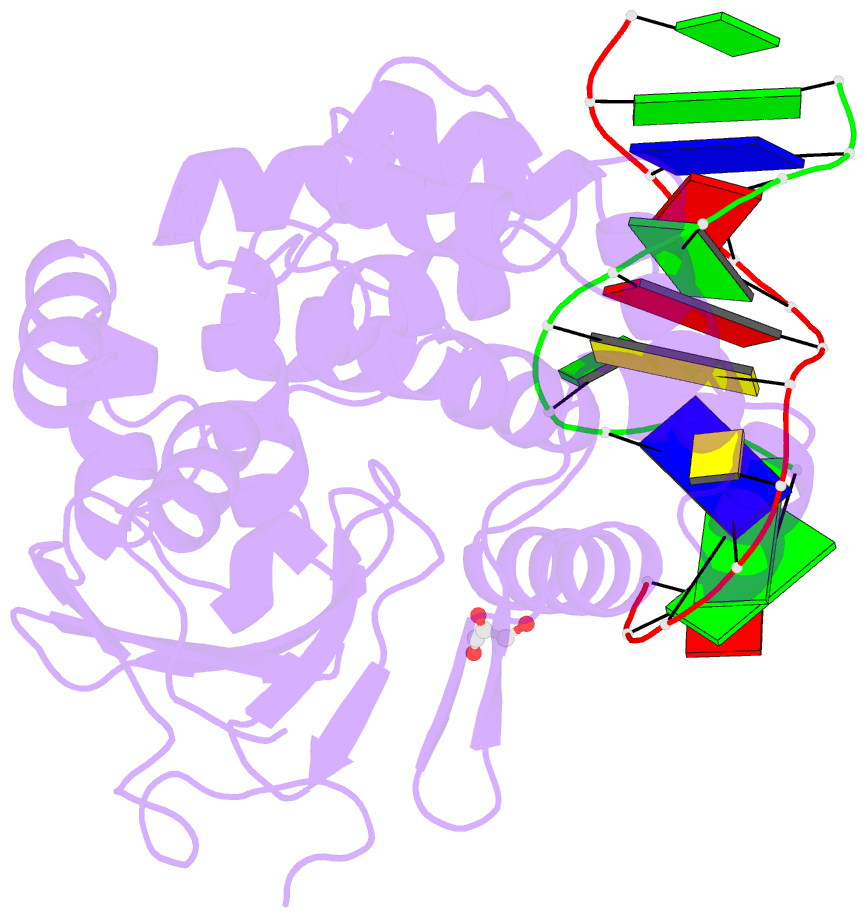
- Structure of hogg1 crosslinked to DNA sampling a normal g adjacent to an oxog
- Reference
- Banerjee A, Verdine GL (2006): "A nucleobase lesion remodels the interaction of its normal neighbor in a DNA glycosylase complex." Proc.Natl.Acad.Sci.Usa, 103, 15020-15025. doi: 10.1073/pnas.0603644103.
- Abstract
- How DNA glycosylases search through millions of base pairs and discriminate between rare sites of damage and otherwise undamaged bases is poorly understood. Even less understood are the details of the structural states arising from DNA glycosylases interacting with undamaged DNA. Recognizing the mutagenic lesion 7,8-dihydro-8-oxoguanine (8-oxoguanine, oxoG) represents an especially formidable challenge, because this oxidized nucleobase differs by only two atoms from its normal counterpart, guanine (G), and buried in the structure of naked B-form DNA, oxoG and G are practically indistinguishable from each other. We have used disulfide cross-linking technology to capture a human oxoG repair protein, 8-oxoguanine DNA glycosylase I (hOGG1) sampling an undamaged G:C base pair located adjacent to an oxoG:C base pair in DNA. The x-ray structure of the trapped complex reveals that the presence of the 8-oxoG drastically changes the local conformation of the extruded G. The extruded but intrahelical state of the G in this structure offers a view of an early intermediate in the base-extrusion pathway.