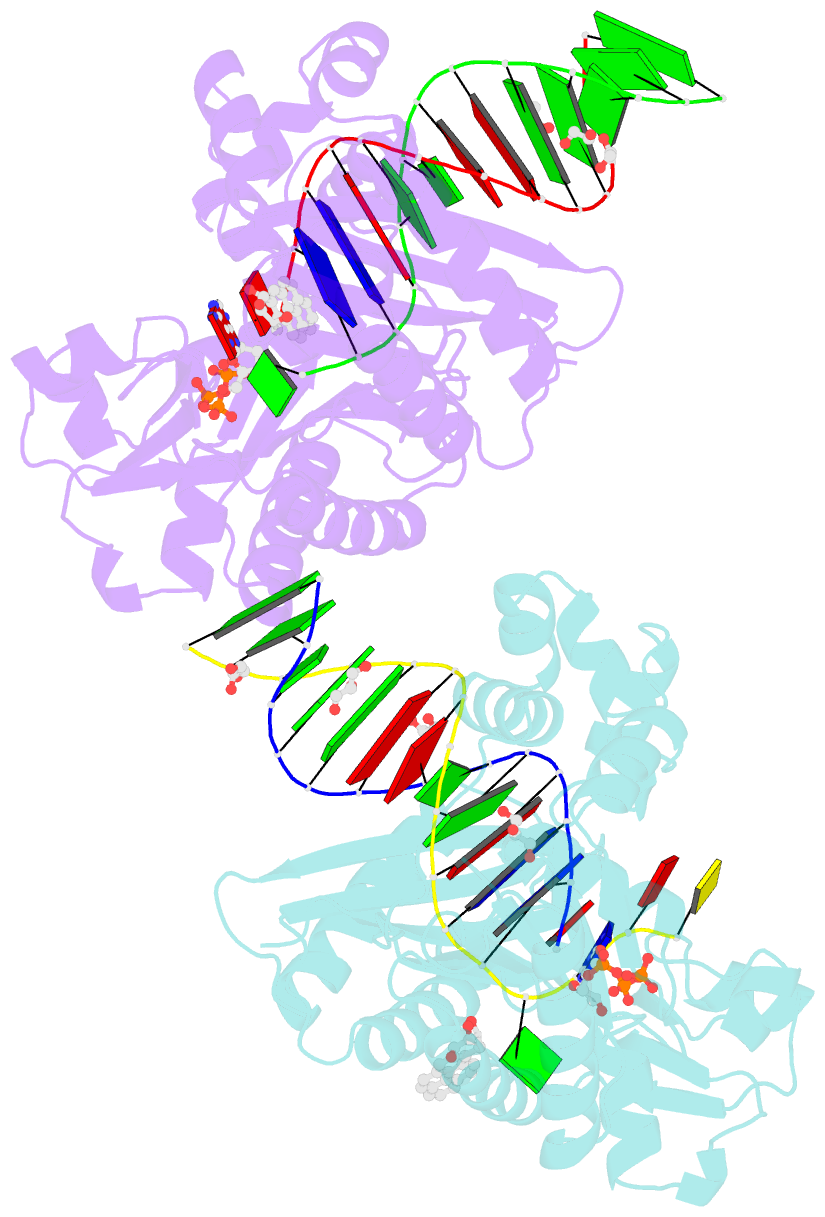
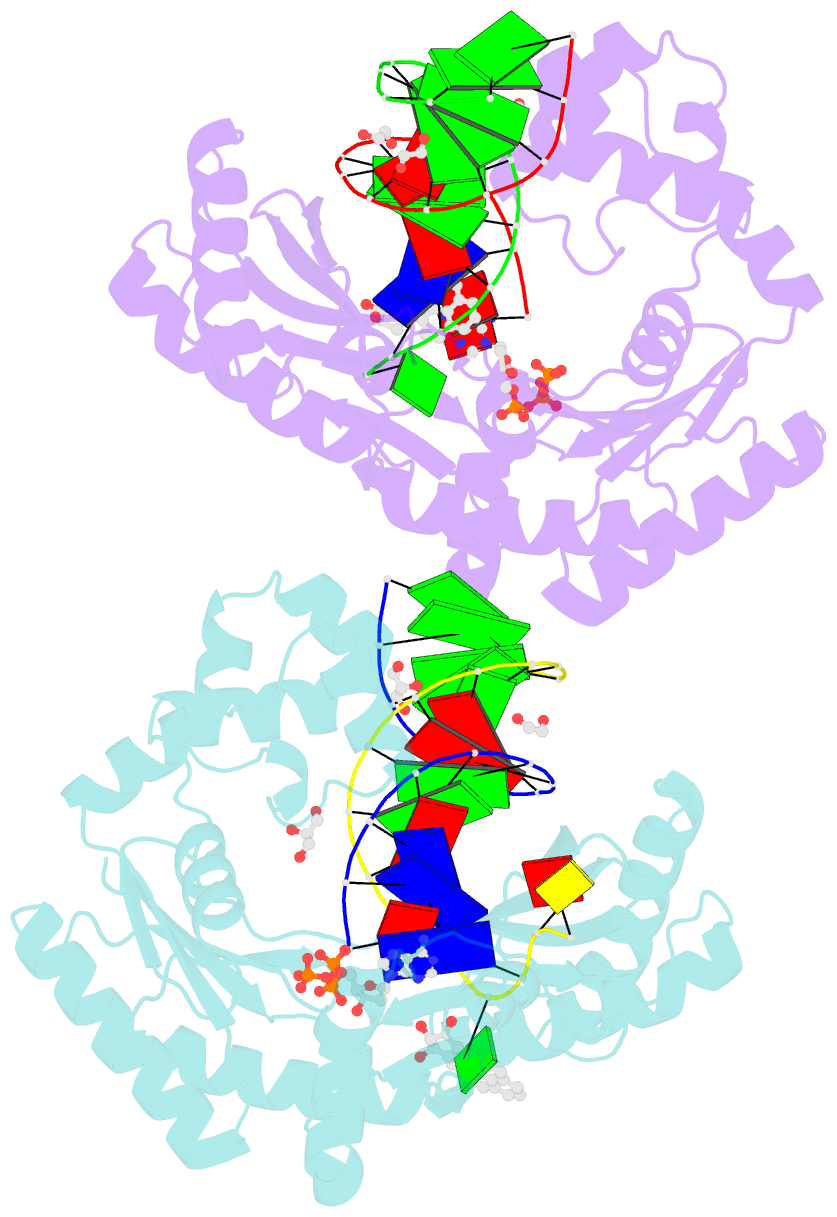
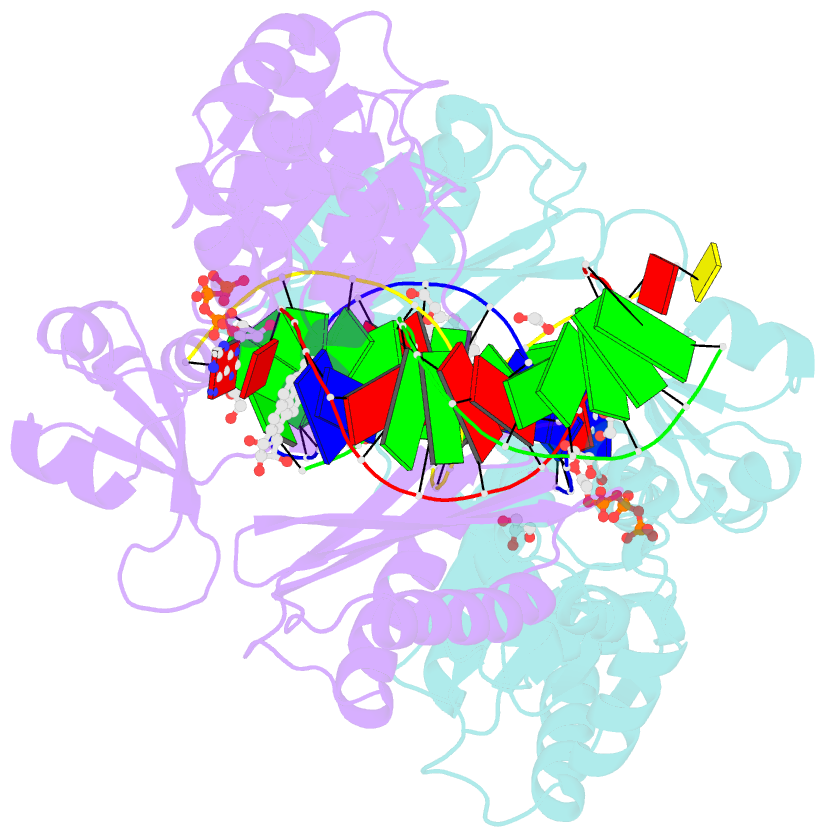
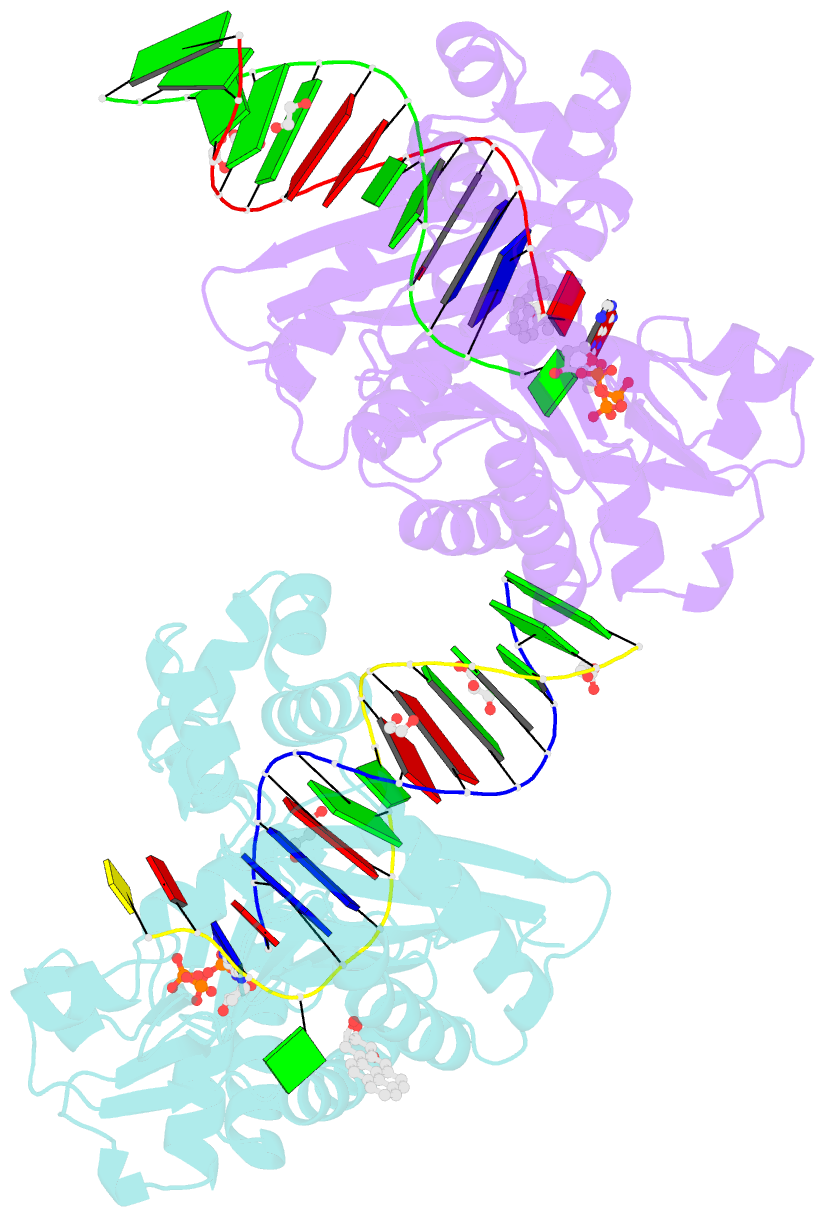
Summary information and primary citation
- PDB-id
- 2ia6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.5 Å)
- Summary
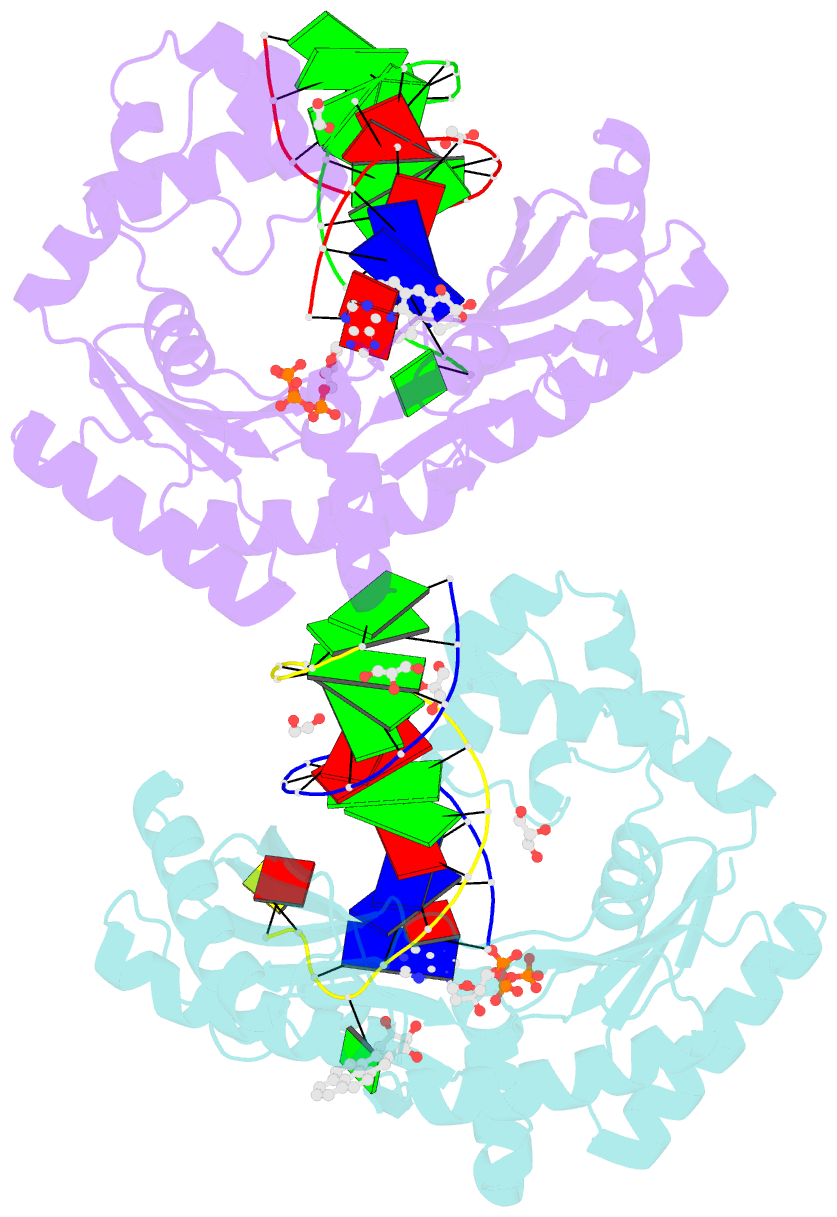
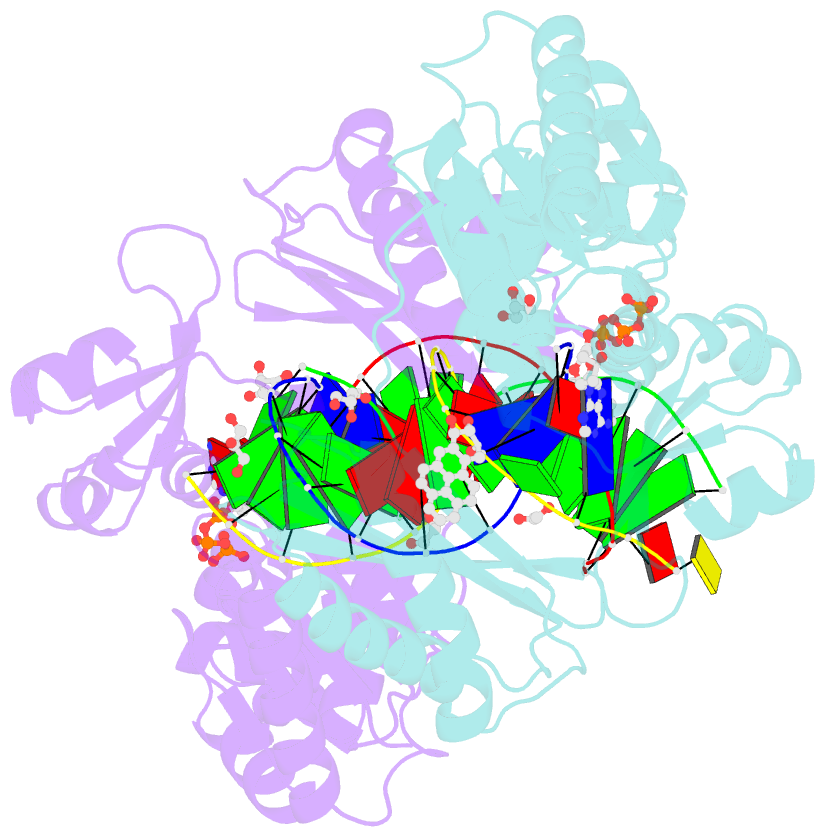
- Bypass of major benzopyrene-dg adduct by y-family DNA polymerase with unique structural gap
- Reference
- Bauer J, Xing G, Yagi H, Sayer JM, Jerina DM, Ling H (2007): "A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment." Proc.Natl.Acad.Sci.Usa, 104, 14905-14910. doi: 10.1073/pnas.0700717104.
- Abstract
- Erroneous replication of lesions in DNA by DNA polymerases leads to elevated mutagenesis. To understand the molecular basis of DNA damage-induced mutagenesis, we have determined the x-ray structures of the Y-family polymerase, Dpo4, in complex with a DNA substrate containing a bulky DNA lesion and incoming nucleotides. The DNA lesion is derived from an environmentally widespread carcinogenic polycyclic aromatic hydrocarbon, benzo[a]pyrene (BP). The potent carcinogen BP is metabolized to diol epoxides that form covalent adducts with cellular DNA. In the present study, the major BP diol epoxide adduct in DNA, BP-N(2)-deoxyguanosine (BP-dG), was placed at a template-primer junction. Three ternary complexes reveal replication blockage, extension past a mismatched lesion, and a -1 frameshift mutation. In the productive structures, the bulky adduct is flipped/looped out of the DNA helix into a structural gap between the little finger and core domains. Sequestering of the hydrophobic BP adduct in this new substrate-binding site permits the DNA to exhibit normal geometry for primer extension. Extrusion of the lesion by template misalignment allows the base 5' to the adduct to serve as the template, resulting in a -1 frameshift. Subsequent strand realignment produces a mismatched base opposite the lesion. These structural observations, in combination with replication and mutagenesis data, suggest a model in which the additional substrate-binding site stabilizes the extrahelical nucleotide for lesion bypass and generation of base substitutions and -1 frameshift mutations.