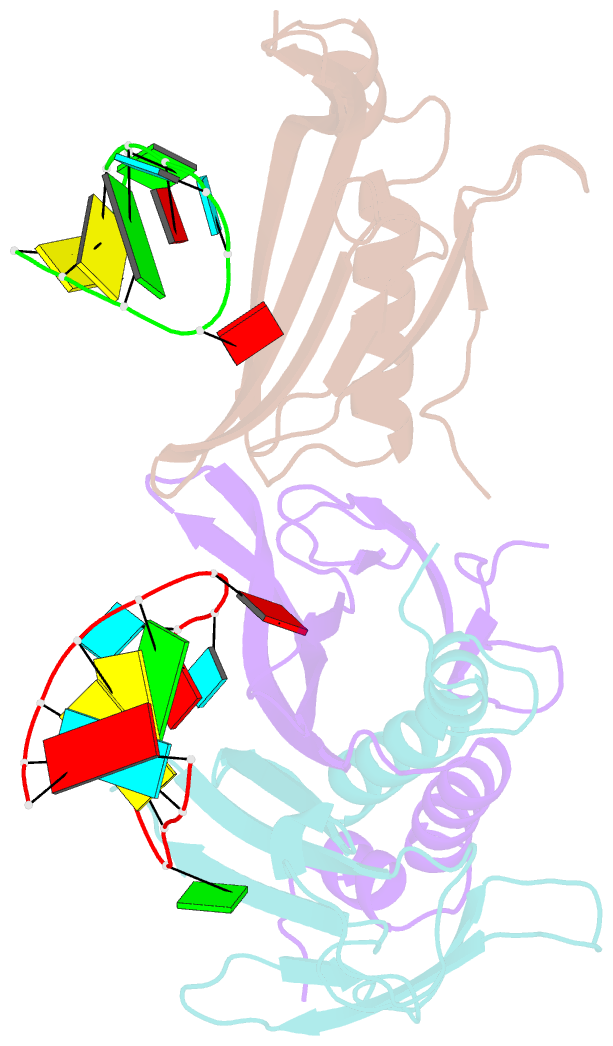
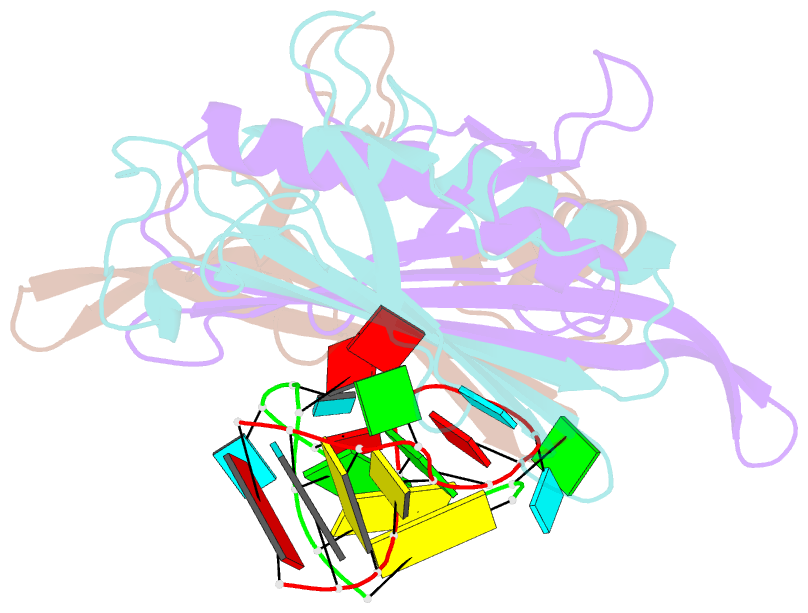
Summary information and primary citation
- PDB-id
- 2izn; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- virus-RNA
- Method
- X-ray (2.56 Å)
- Summary
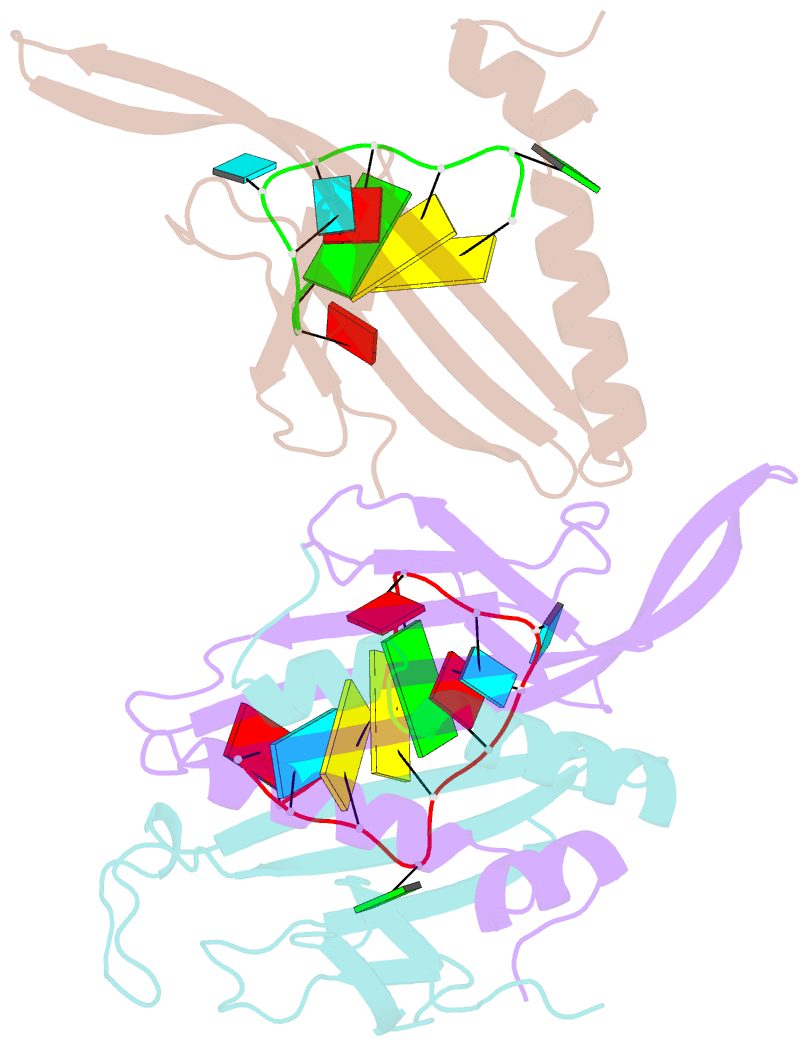
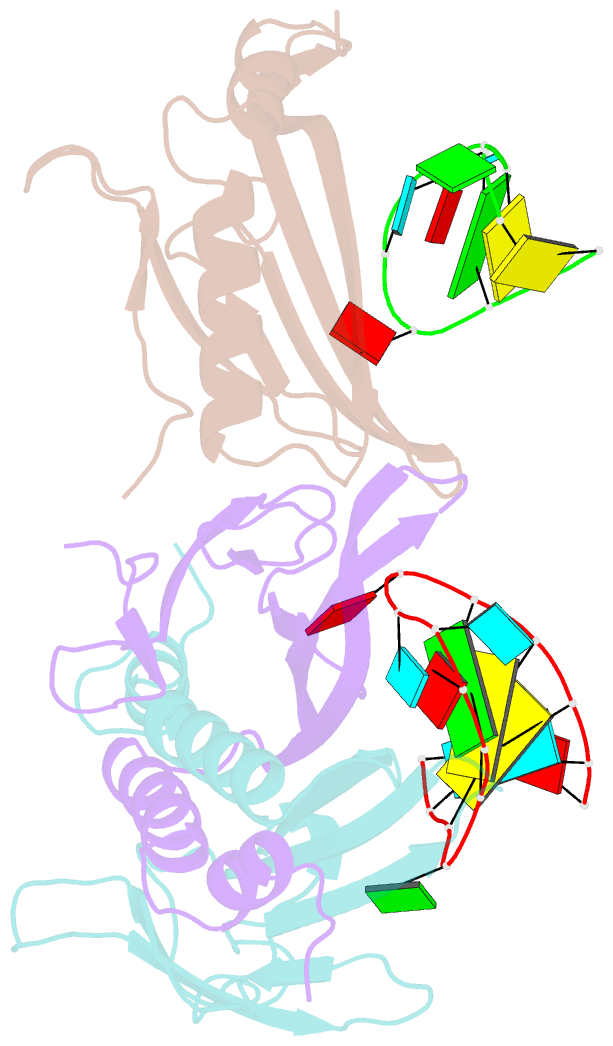
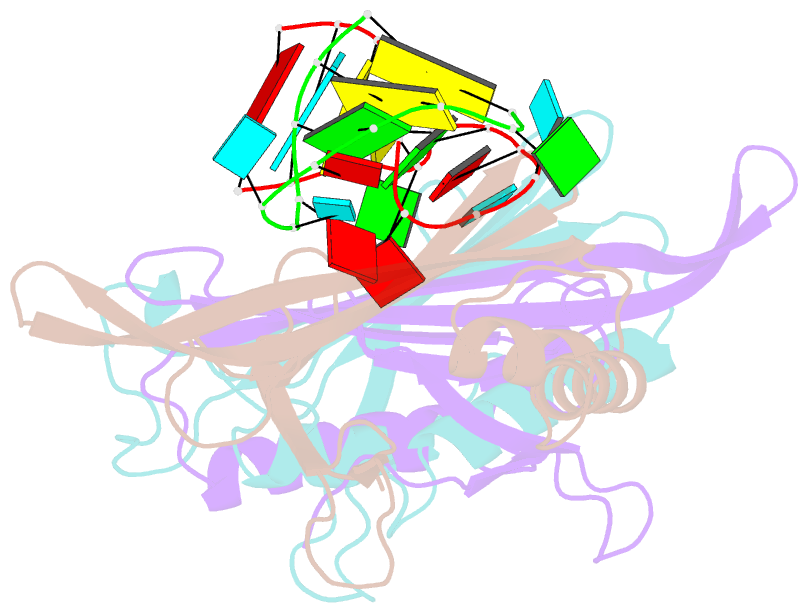
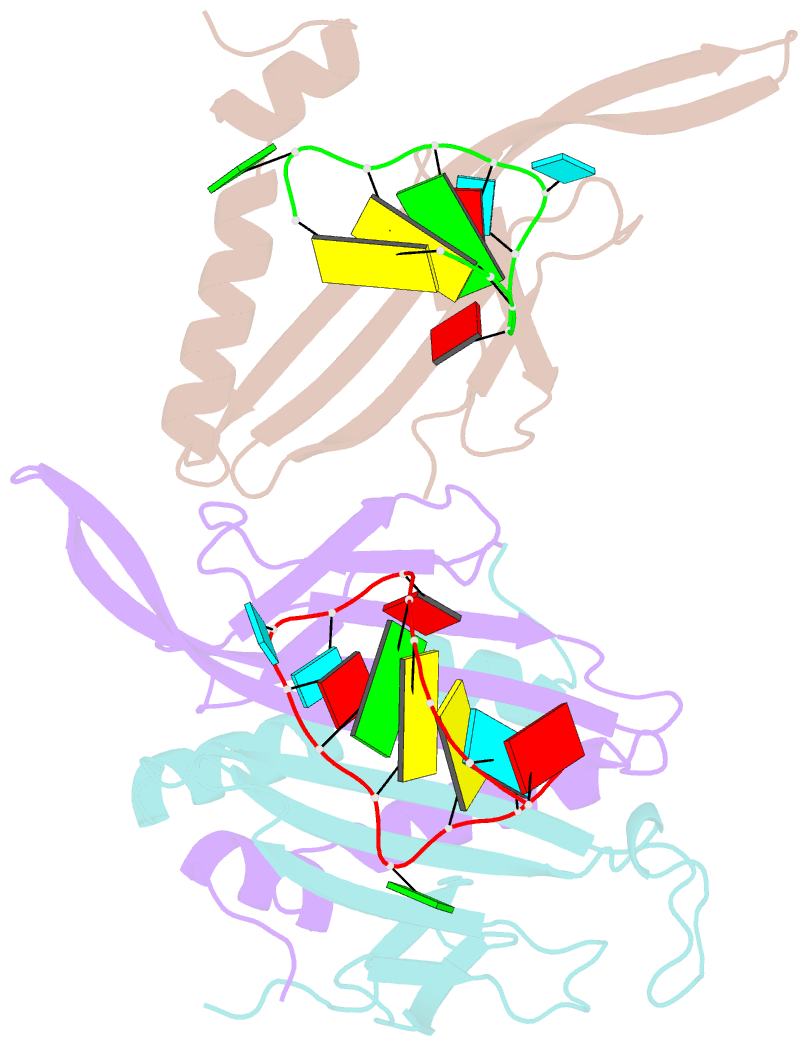
- Ms2-RNA hairpin (g-10) complex
- Reference
- Helgstrand C, Grahn E, Moss T, Stonehouse NJ, Tars K, Stockley PG, Liljas L (2002): "Investigating the Structural Basis of Purine Specificity in the Structures of MS2 Coat Protein RNA Translational Operator Complexes." Nucleic Acids Res., 30, 2678. doi: 10.1093/NAR/GKF371.
- Abstract
- We have determined the structures of complexes between the phage MS2 coat protein and variants of the replicase translational operator in order to explore the sequence specificity of the RNA-protein interaction. The 19-nt RNA hairpins studied have substitutions at two positions that have been shown to be important for specific binding. At one of these positions, -10, which is a bulged adenosine (A) in the stem of the wild-type operator hairpin, substitutions were made with guanosine (G), cytidine (C) and two non-native bases, 2-aminopurine (2AP) and inosine (I). At the other position, -7 in the hairpin loop, the native adenine was substituted with a cytidine. Of these, only the G-10, C-10 and C-7 variants showed interpretable density for the RNA hairpin. In spite of large differences in binding affinities, the structures of the variant complexes are very similar to the wild-type operator complex. For G-10 substitutions in hairpin variants that can form bulges at alternative places in the stem, the binding affinity is low and a partly disordered conformation is seen in the electron density maps. The affinity is similar to that of wild-type when the base pairs adjacent to the bulged nucleotide are selected to avoid alternative conformations. Both purines bind in a very similar way in a pocket in the protein. In the C-10 variant, which has very low affinity, the cytidine is partly inserted in the protein pocket rather than intercalated in the RNA stem. Substitution of the wild-type adenosine at position -7 by pyrimidines gives strongly reduced affinities, but the structure of the C-7 complex shows that the base occupies the same position as the A-7 in the wild-type RNA. It is stacked in the RNA and makes no direct contact with the protein.