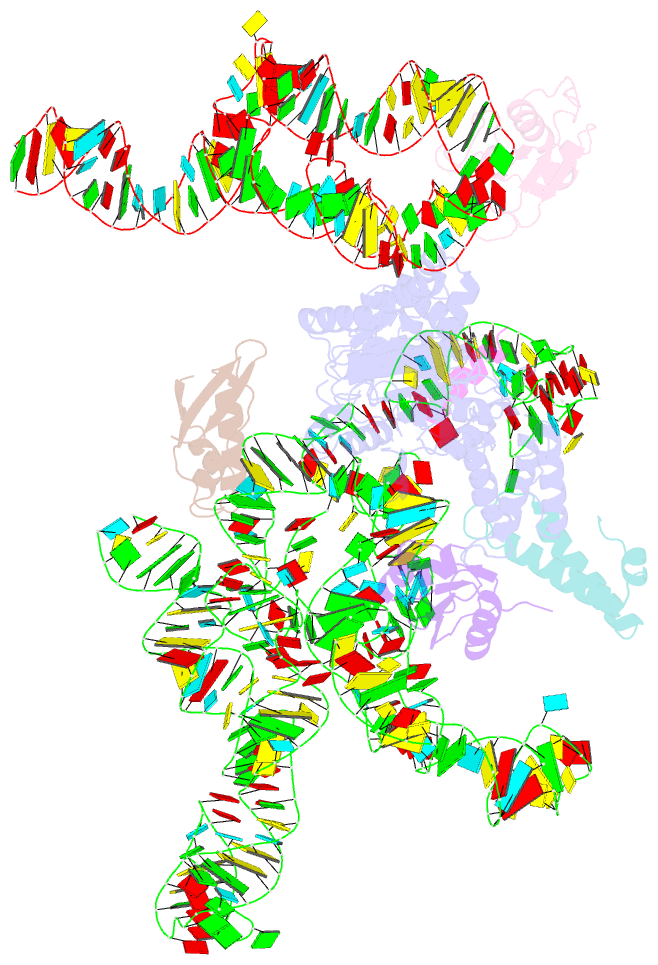
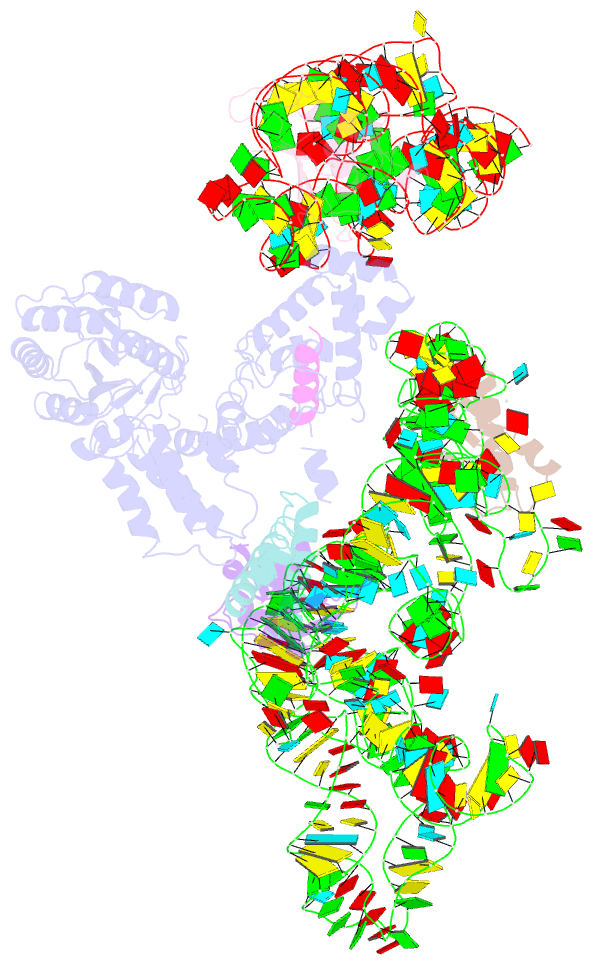
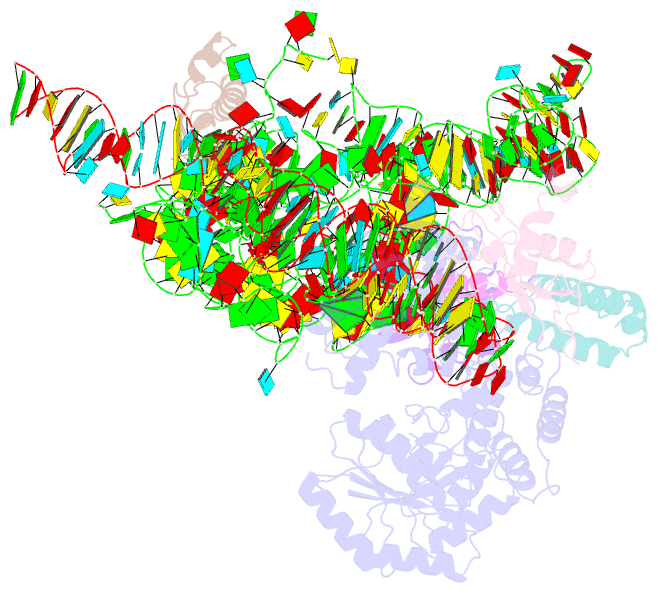
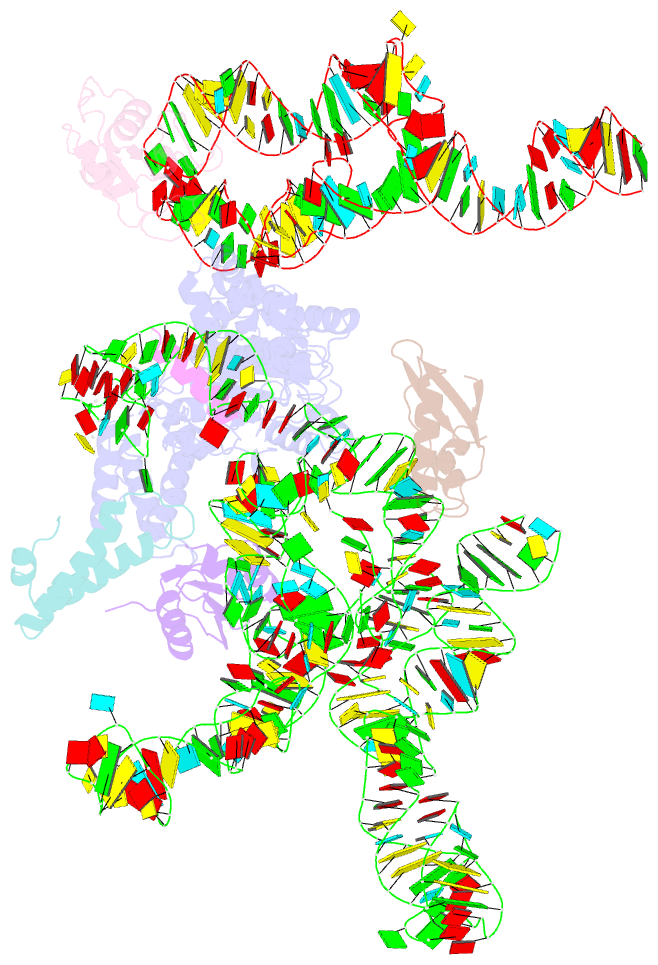
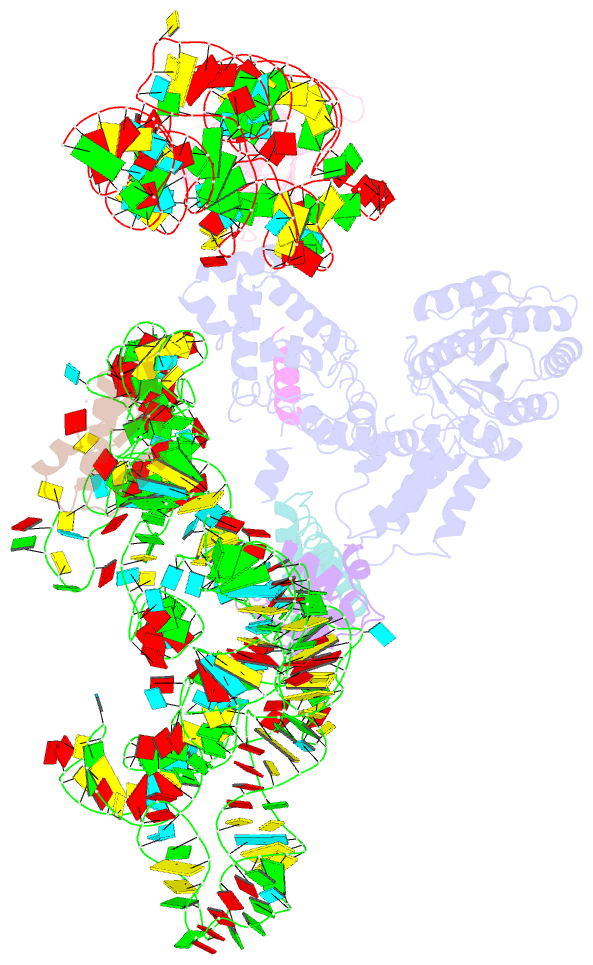
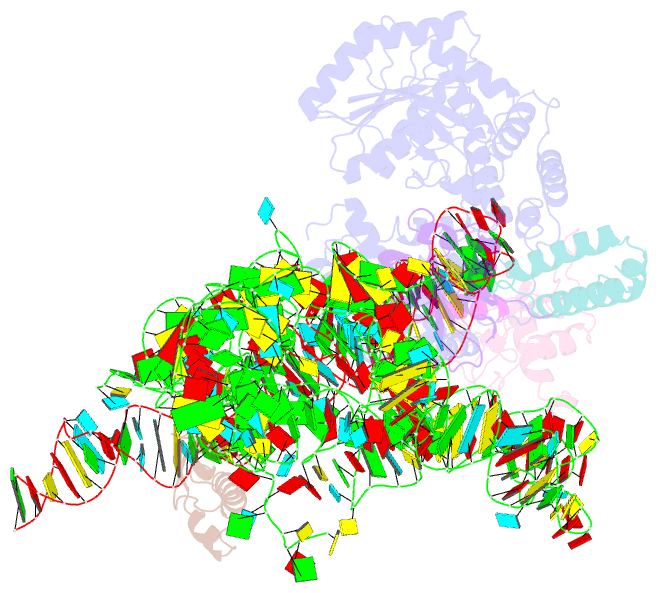
Summary information and primary citation
- PDB-id
- 2j37; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- cryo-EM (8.7 Å)
- Summary
- Model of mammalian srp bound to 80s rncs
- Reference
- Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R (2006): "Following the signal sequence from ribosomal tunnel exit to signal recognition particle." Nature, 444, 507-511. doi: 10.1038/nature05326.
- Abstract
- Membrane and secretory proteins can be co-translationally inserted into or translocated across the membrane. This process is dependent on signal sequence recognition on the ribosome by the signal recognition particle (SRP), which results in targeting of the ribosome-nascent-chain complex to the protein-conducting channel at the membrane. Here we present an ensemble of structures at subnanometre resolution, revealing the signal sequence both at the ribosomal tunnel exit and in the bacterial and eukaryotic ribosome-SRP complexes. Molecular details of signal sequence interaction in both prokaryotic and eukaryotic complexes were obtained by fitting high-resolution molecular models. The signal sequence is presented at the ribosomal tunnel exit in an exposed position ready for accommodation in the hydrophobic groove of the rearranged SRP54 M domain. Upon ribosome binding, the SRP54 NG domain also undergoes a conformational rearrangement, priming it for the subsequent docking reaction with the NG domain of the SRP receptor. These findings provide the structural basis for improving our understanding of the early steps of co-translational protein sorting.