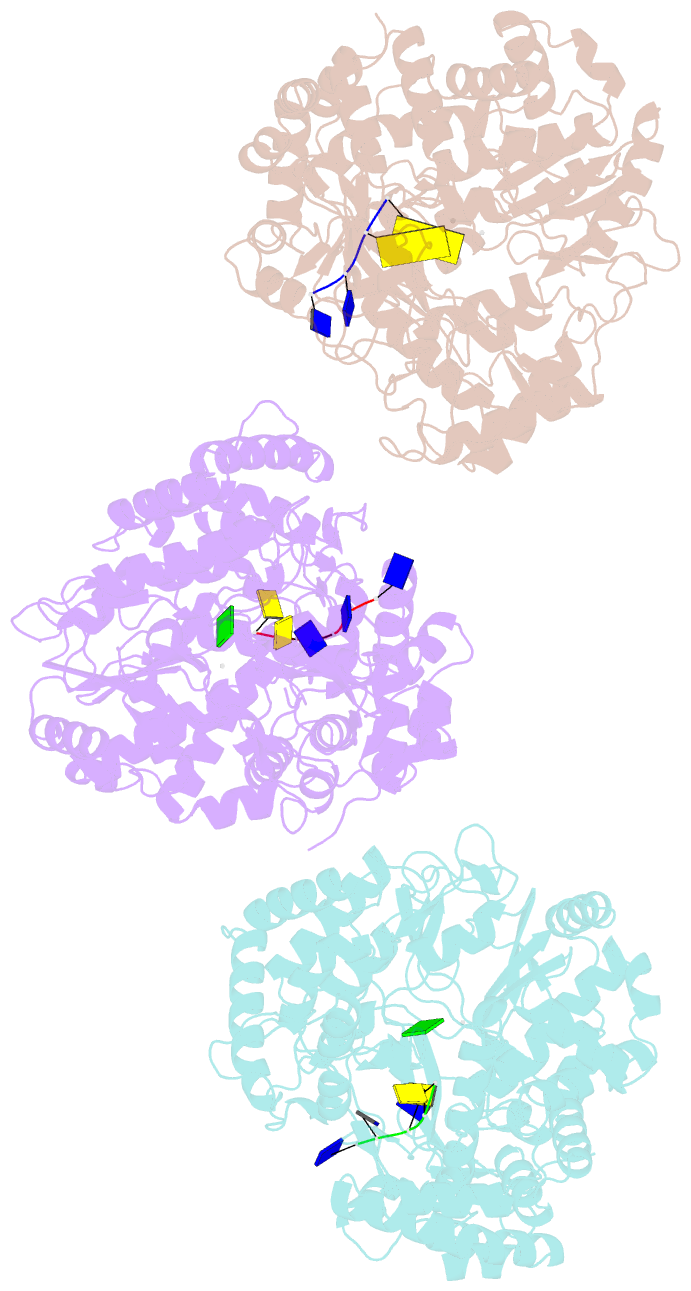
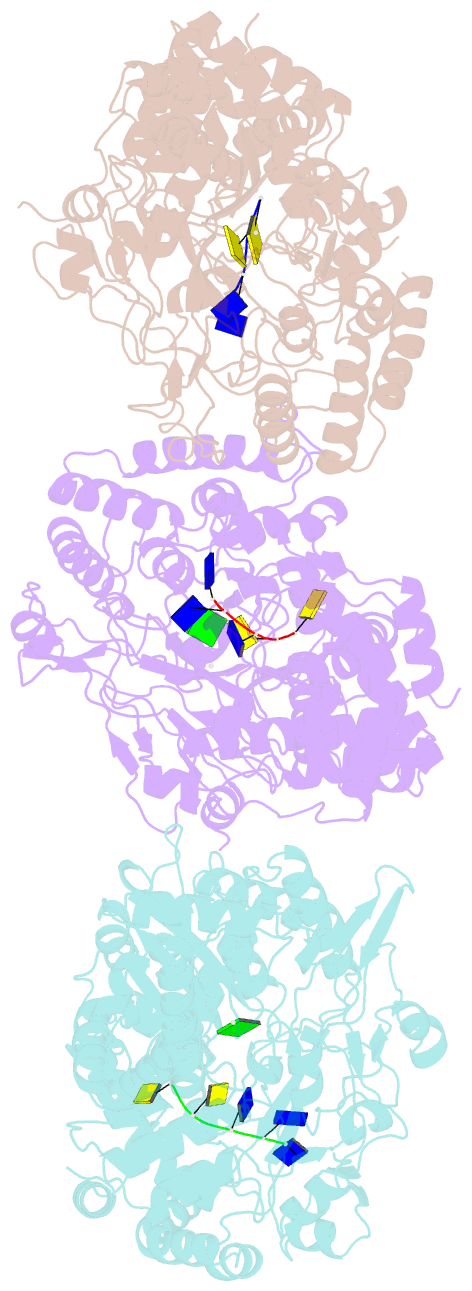
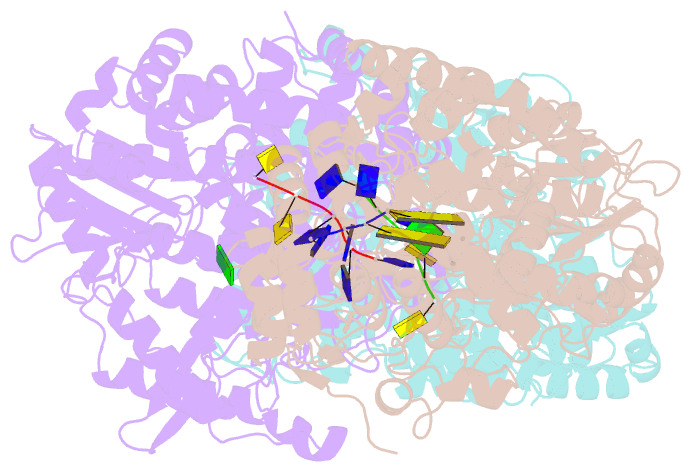
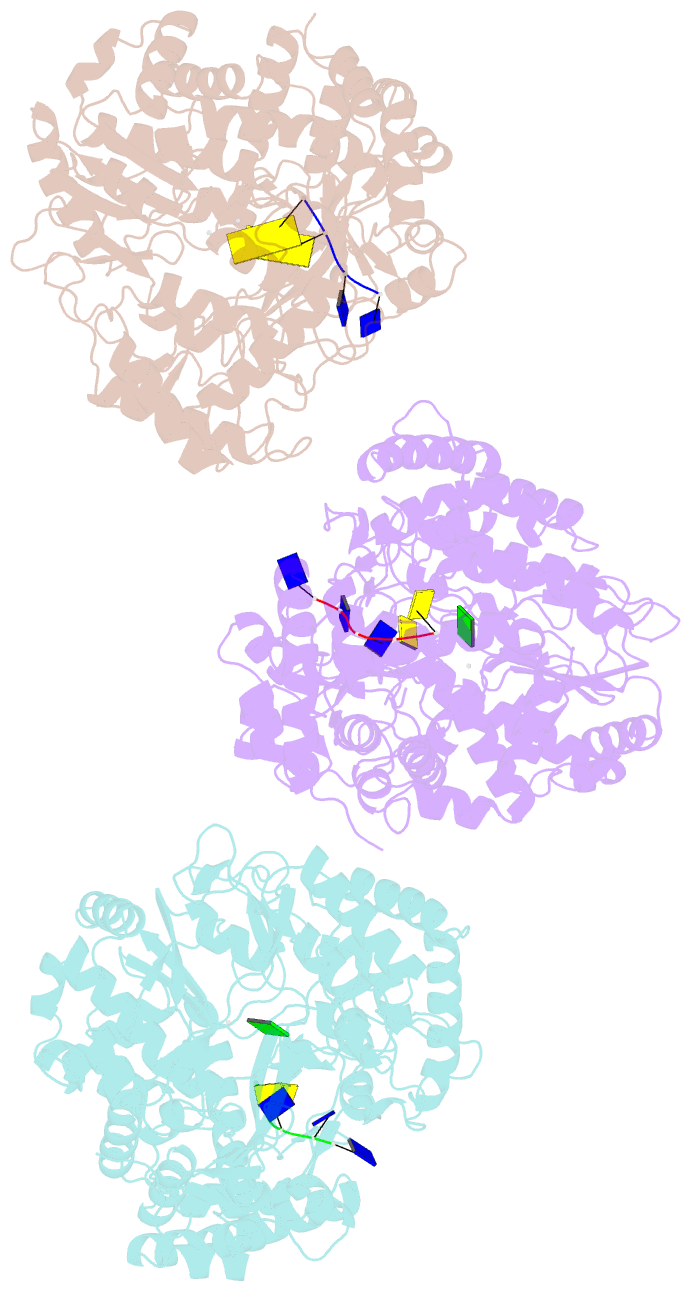
Summary information and primary citation
- PDB-id
- 2jlg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase
- Method
- X-ray (2.8 Å)
- Summary
- Structural explanation for the role of mn in the activity of phi6 RNA-dependent RNA polymerase
- Reference
- Poranen MM, Salgado PS, Koivunen MRL, Wright S, Bamford DH, Stuart DI, Grimes JM (2008): "Structural Explanation for the Role of Mn2+ in the Activity of {Phi}6 RNA-Dependent RNA Polymerase." Nucleic Acids Res., 36, 6633. doi: 10.1093/NAR/GKN632.
- Abstract
- The biological role of manganese (Mn(2+)) has been a long-standing puzzle, since at low concentrations it activates several polymerases whilst at higher concentrations it inhibits. Viral RNA polymerases possess a common architecture, reminiscent of a closed right hand. The RNA-dependent RNA polymerase (RdRp) of bacteriophage 6 is one of the best understood examples of this important class of polymerases. We have probed the role of Mn(2+) by biochemical, biophysical and structural analyses of the wild-type enzyme and of a mutant form with an altered Mn(2+)-binding site (E491 to Q). The E491Q mutant has much reduced affinity for Mn(2+), reduced RNA binding and a compromised elongation rate. Loss of Mn(2+) binding structurally stabilizes the enzyme. These data and a re-examination of the structures of other viral RNA polymerases clarify the role of manganese in the activation of polymerization: Mn(2+) coordination of a catalytic aspartate is necessary to allow the active site to properly engage with the triphosphates of the incoming NTPs. The structural flexibility caused by Mn(2+) is also important for the enzyme dynamics, explaining the requirement for manganese throughout RNA polymerization.