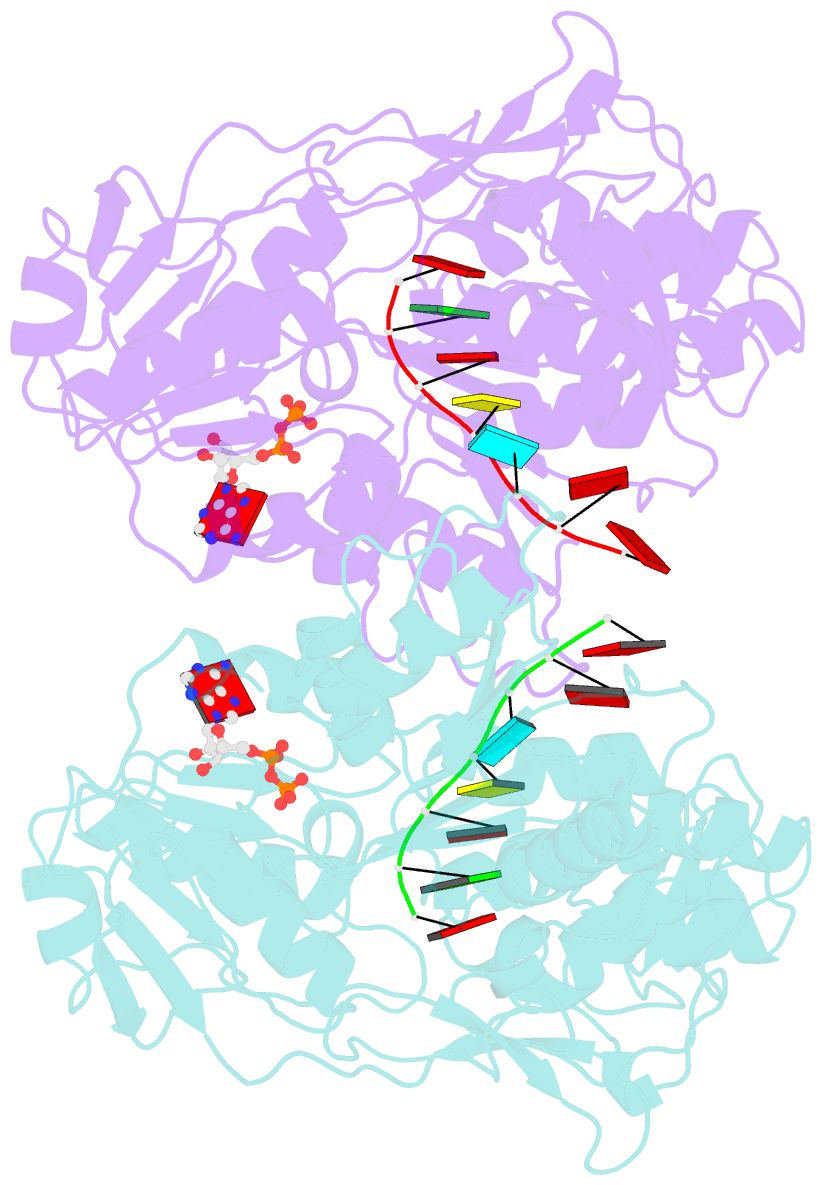
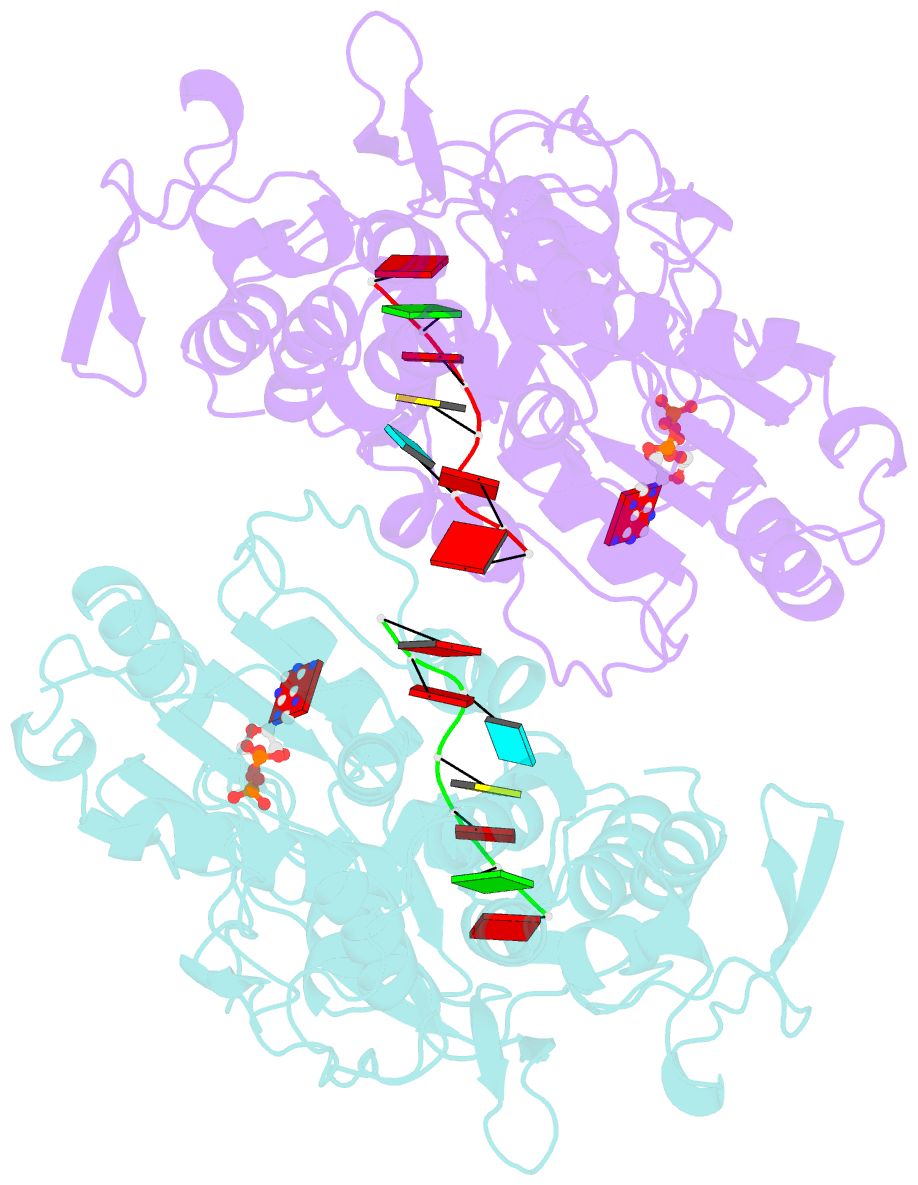
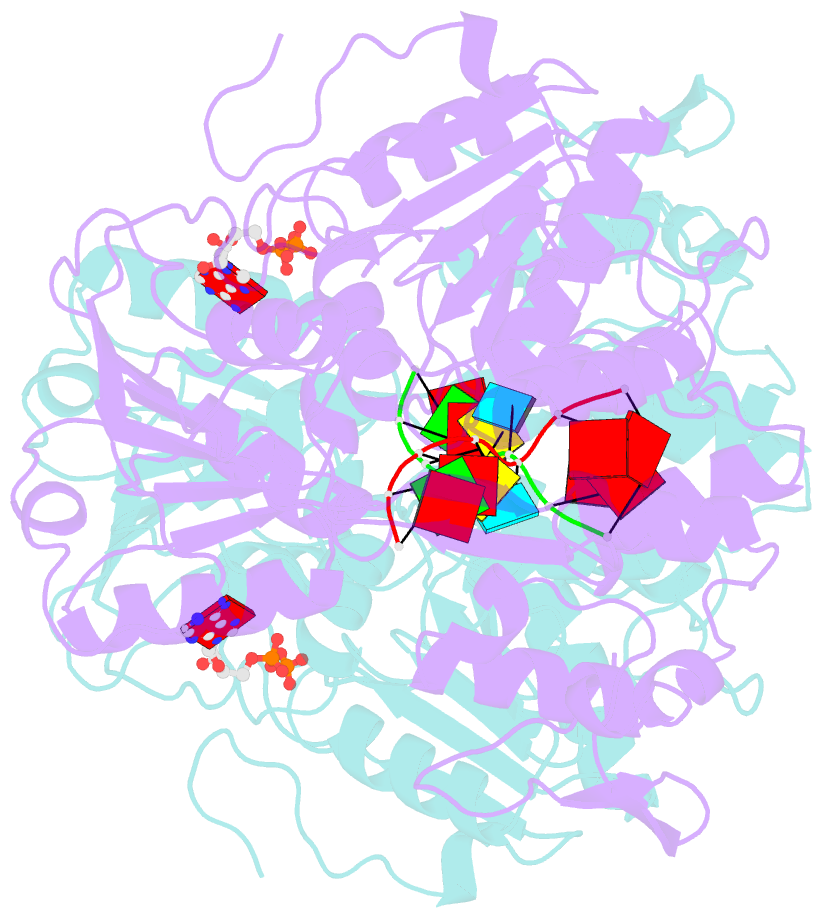
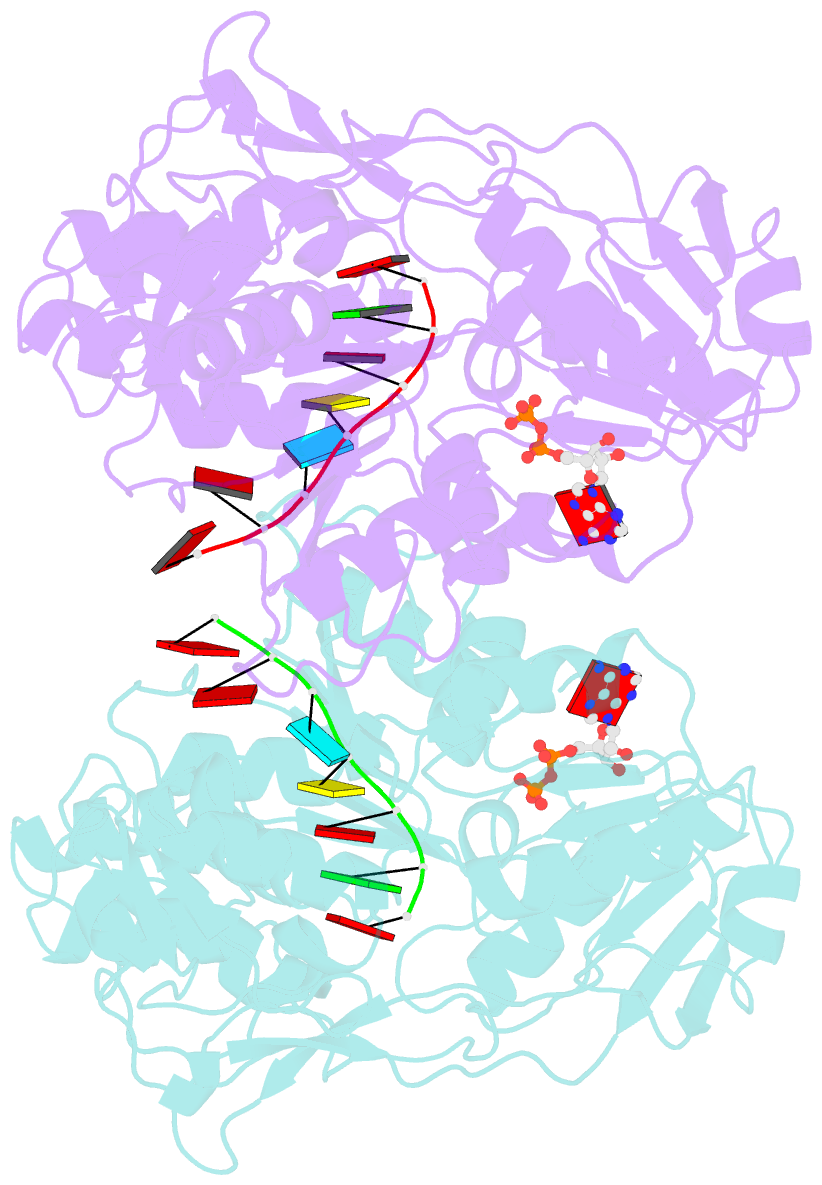
Summary information and primary citation
- PDB-id
- 2jlx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.2 Å)
- Summary
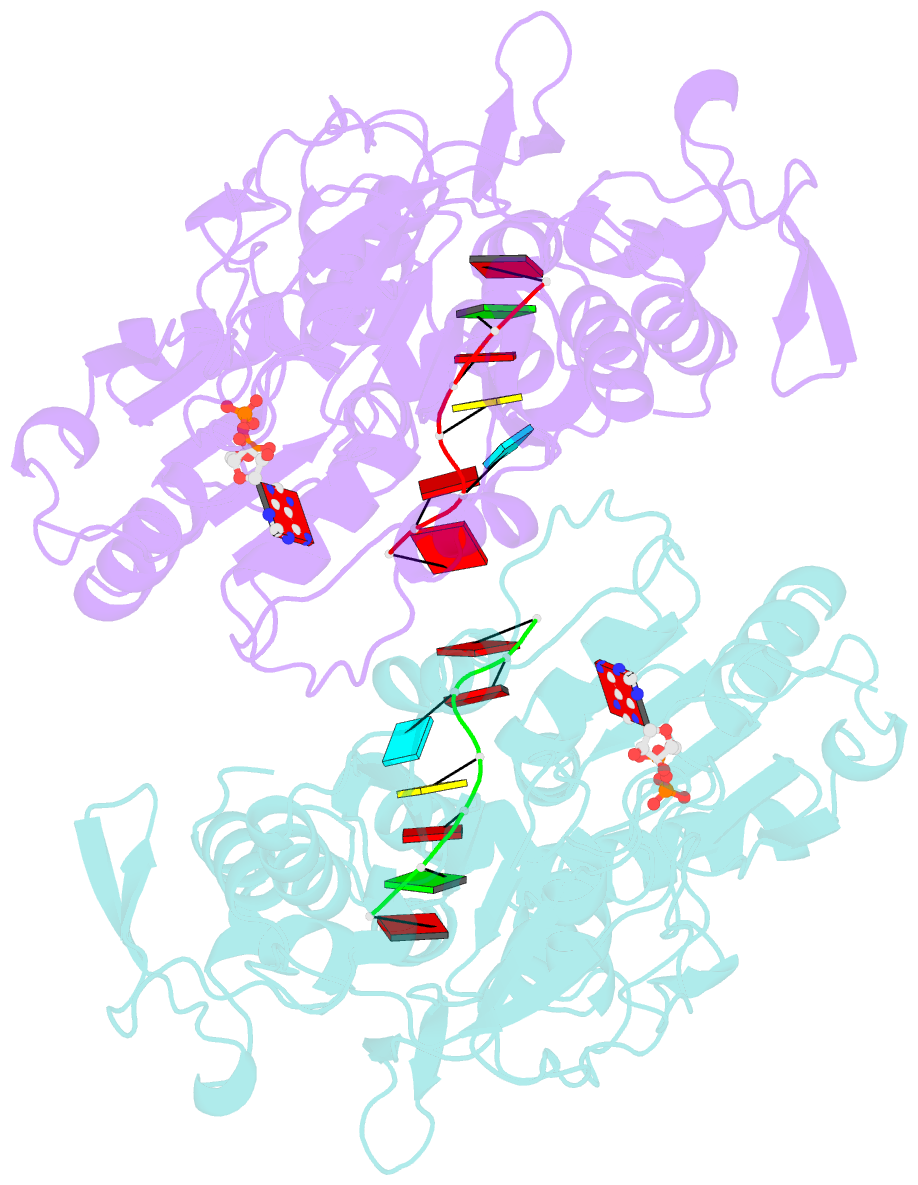
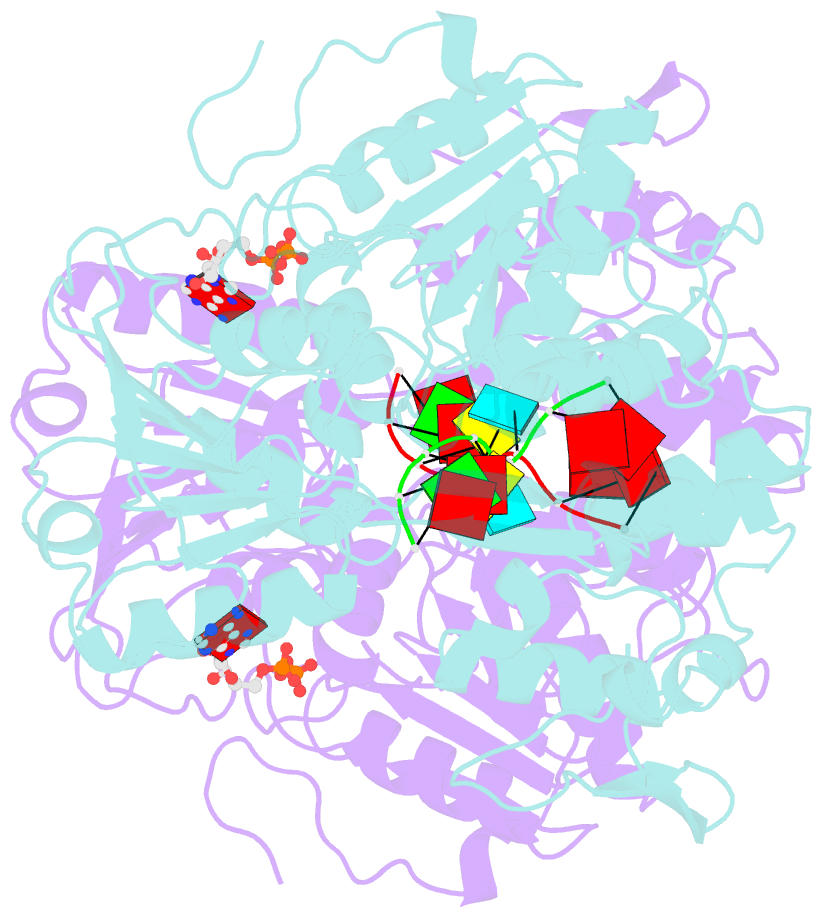
- Dengue virus 4 ns3 helicase in complex with ssrna and adp-vanadate
- Reference
- Luo DH, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, Yeong SS, Wang CH, Lim SP, Strongin A, Vasudevan SG, Lescar J (2008): "Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein." Embo J., 27, 3209. doi: 10.1038/EMBOJ.2008.232.
- Abstract
- Together with the NS5 polymerase, the NS3 helicase has a pivotal function in flavivirus RNA replication and constitutes an important drug target. We captured the dengue virus NS3 helicase at several stages along the catalytic pathway including bound to single-stranded (ss) RNA, to an ATP analogue, to a transition-state analogue and to ATP hydrolysis products. RNA recognition appears largely sequence independent in a way remarkably similar to eukaryotic DEAD box proteins Vasa and eIF4AIII. On ssRNA binding, the NS3 enzyme switches to a catalytic-competent state imparted by an inward movement of the P-loop, interdomain closure and a change in the divalent metal coordination shell, providing a structural basis for RNA-stimulated ATP hydrolysis. These structures demonstrate for the first time large quaternary changes in the flaviviridae helicase, identify the catalytic water molecule and point to a beta-hairpin that protrudes from subdomain 2, as a critical element for dsRNA unwinding. They also suggest how NS3 could exert an effect as an RNA-anchoring device and thus participate both in flavivirus RNA replication and assembly.