Summary information and primary citation
- PDB-id
- 2jq7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome-antibiotic
- Method
- NMR
- Summary
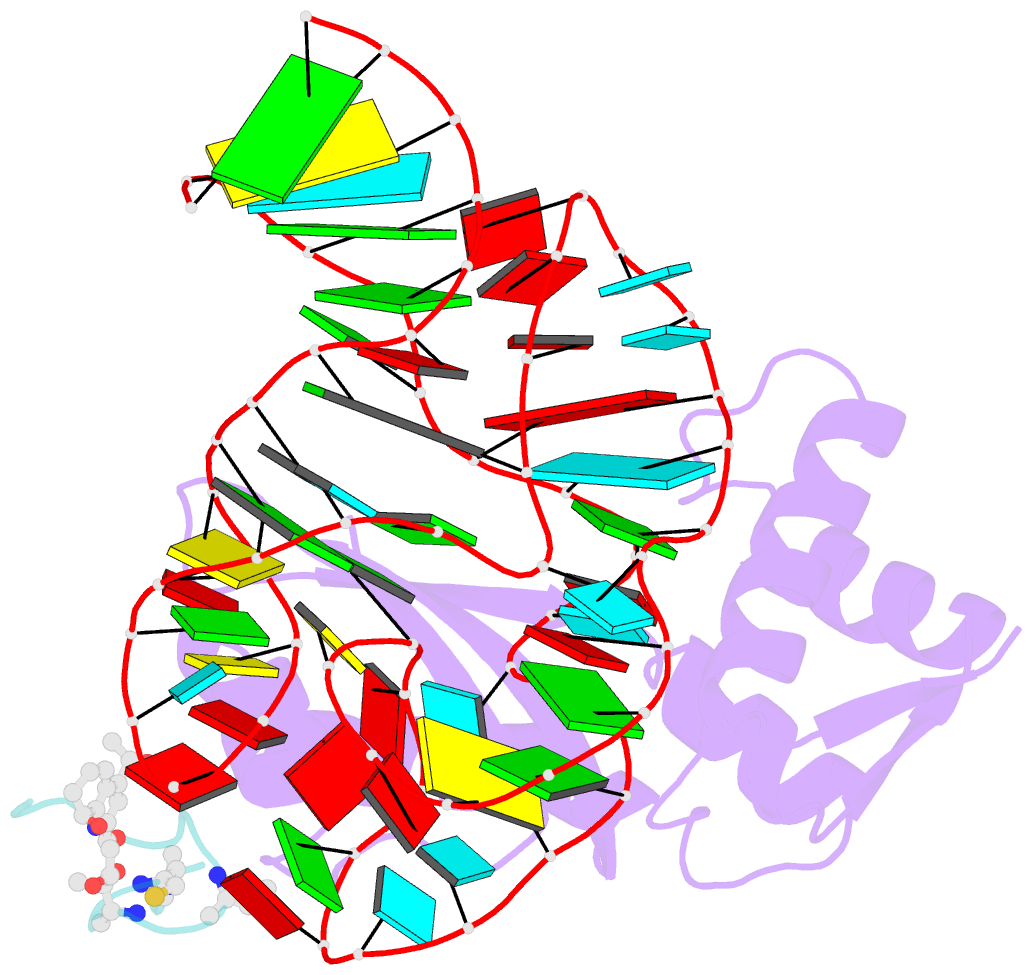
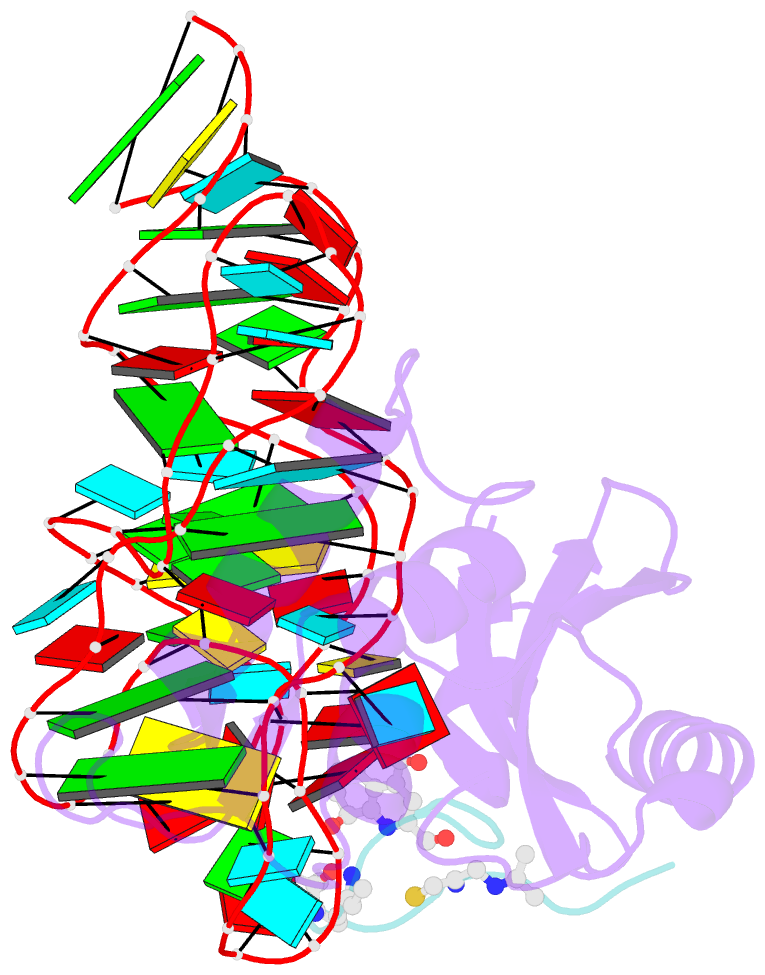
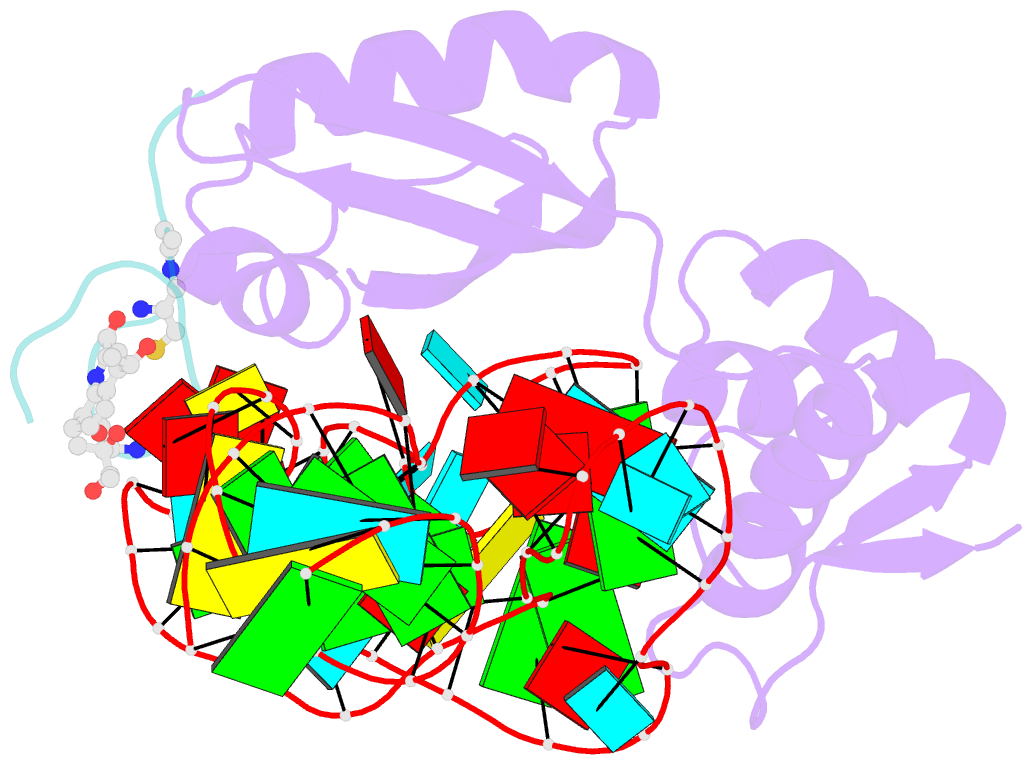
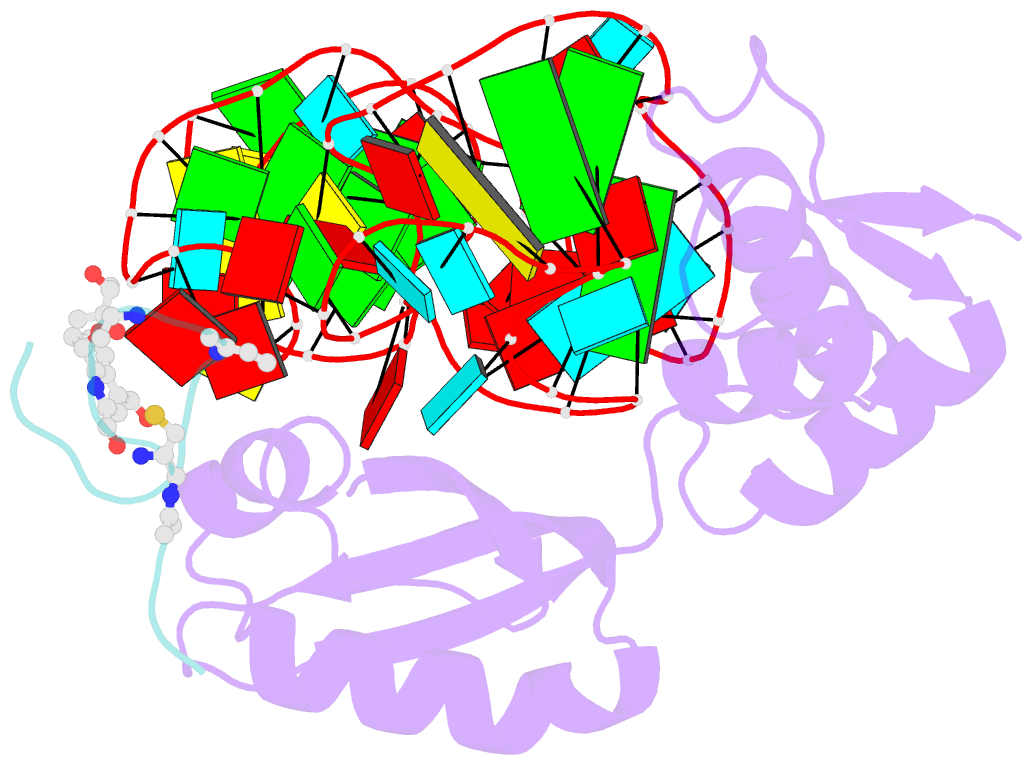
- Model for thiostrepton binding to the ribosomal l11-RNA
- Reference
- Jonker HRA, Ilin S, Grimm SK, Woehnert J, Schwalbe H (2007): "L11 Domain Rearrangement Upon Binding to RNA and Thiostrepton Studied by NMR Spectroscopy." Nucleic Acids Res., 35, 441. doi: 10.1093/NAR/GKL1066.
- Abstract
- Ribosomal proteins are assumed to stabilize specific RNA structures and promote compact folding of the large rRNA. The conformational dynamics of the protein between the bound and unbound state play an important role in the binding process. We have studied those dynamical changes in detail for the highly conserved complex between the ribosomal protein L11 and the GTPase region of 23S rRNA. The RNA domain is compactly folded into a well defined tertiary structure, which is further stabilized by the association with the C-terminal domain of the L11 protein (L11(ctd)). In addition, the N-terminal domain of L11 (L11(ntd)) is implicated in the binding of the natural thiazole antibiotic thiostrepton, which disrupts the elongation factor function. We have studied the conformation of the ribosomal protein and its dynamics by NMR in the unbound state, the RNA bound state and in the ternary complex with the RNA and thiostrepton. Our data reveal a rearrangement of the L11(ntd), placing it closer to the RNA after binding of thiostrepton, which may prevent binding of elongation factors. We propose a model for the ternary L11-RNA-thiostrepton complex that is additionally based on interaction data and conformational information of the L11 protein. The model is consistent with earlier findings and provides an explanation for the role of L11(ntd) in elongation factor binding.