Summary information and primary citation
- PDB-id
- 2kdz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- NMR
- Summary
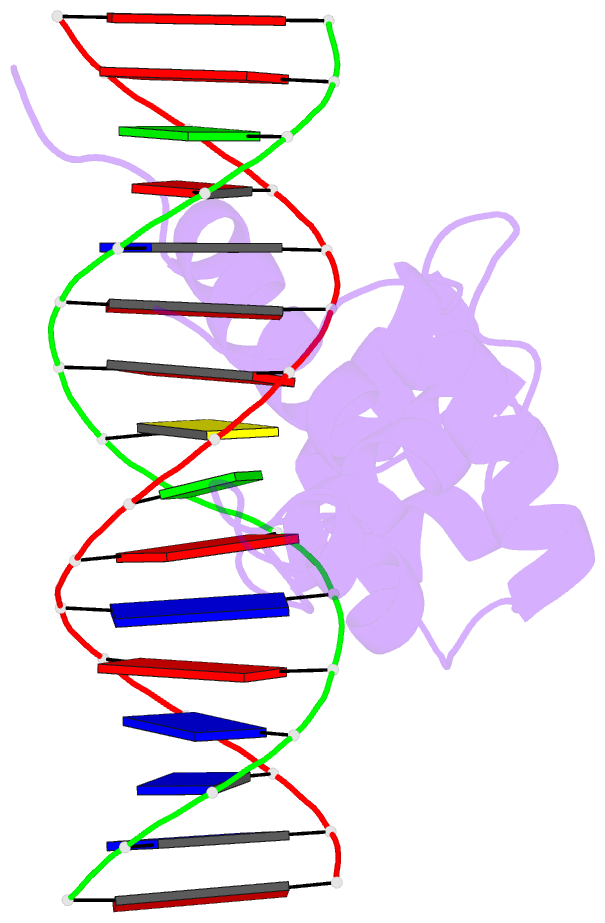
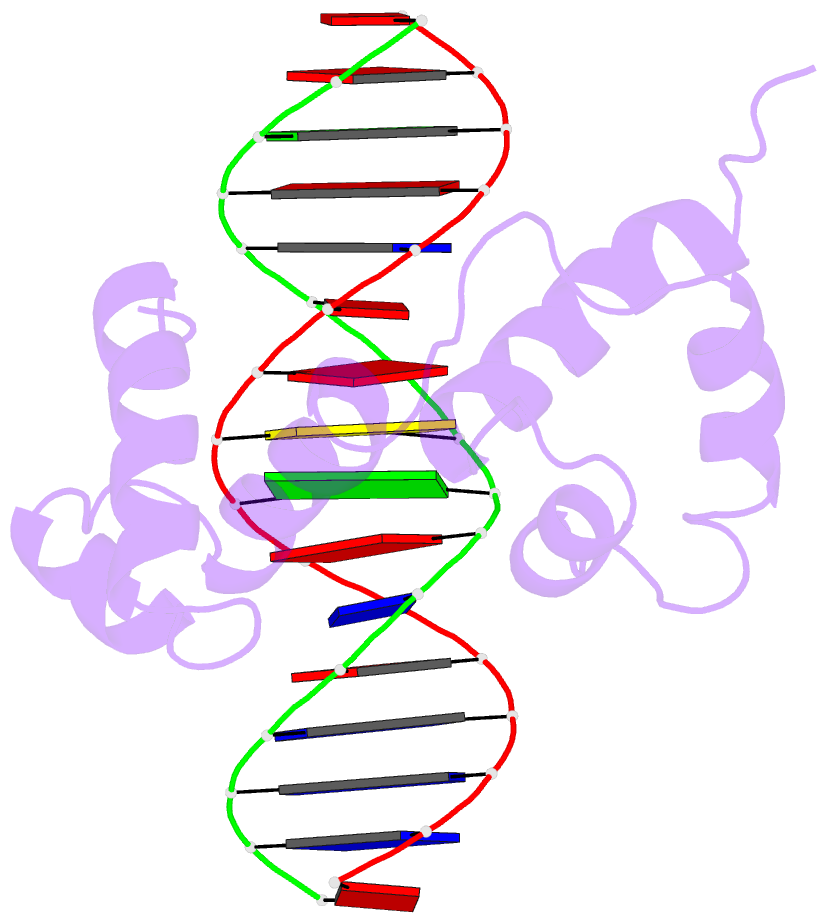
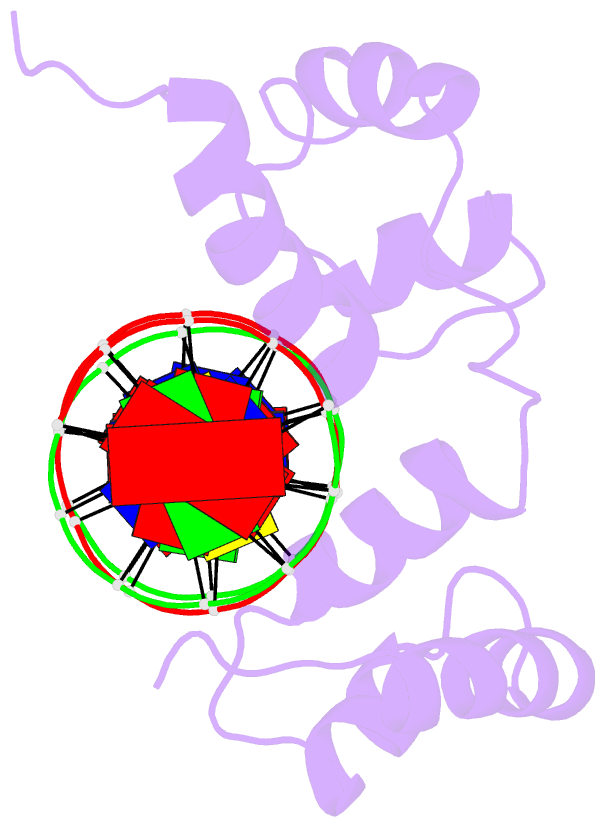
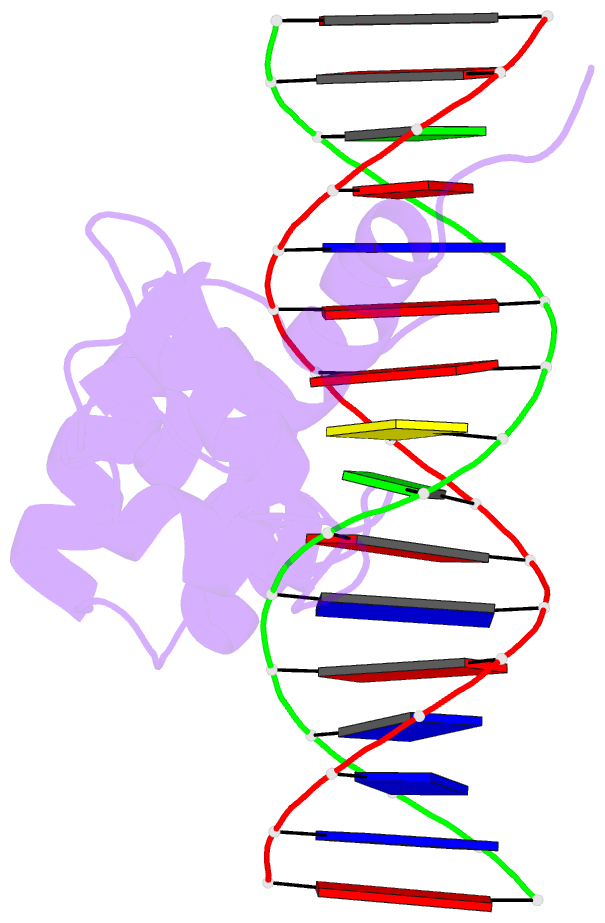
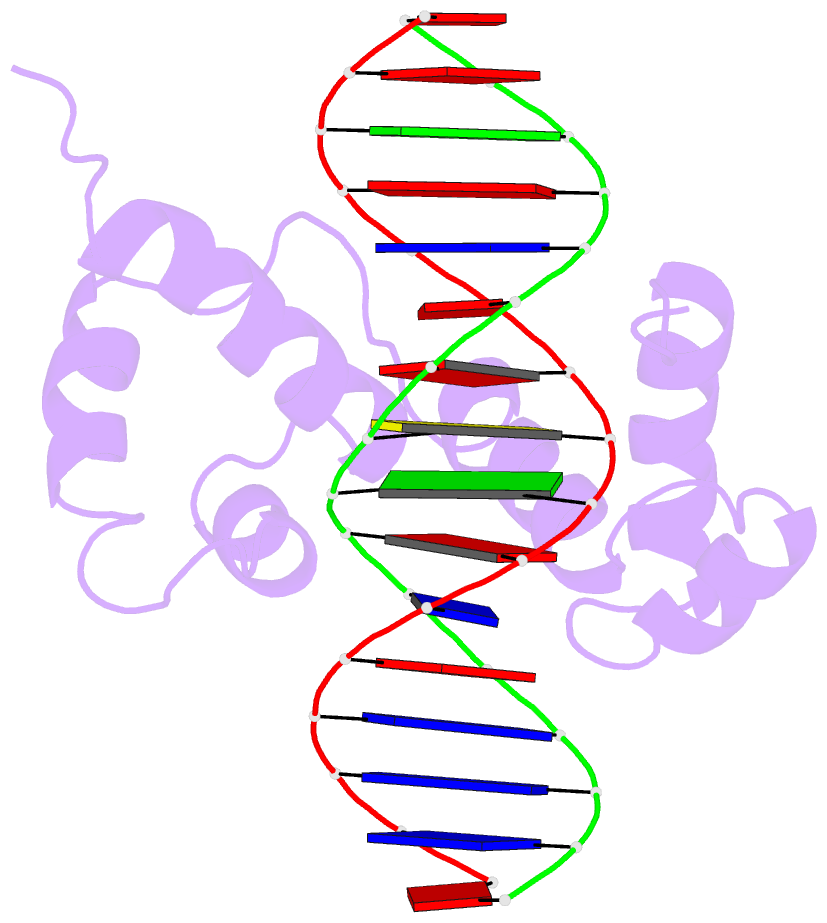
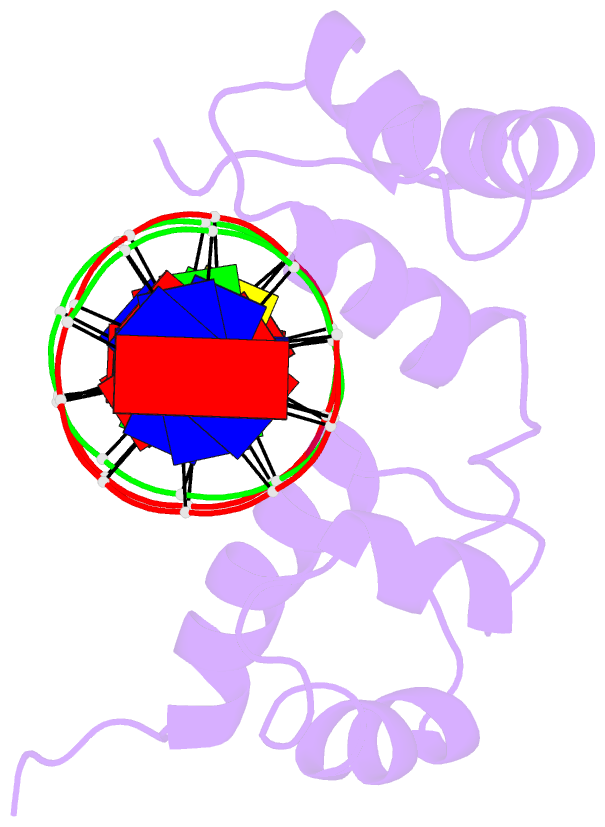
- Structure of the r2r3 DNA binding domain of myb1 protein from protozoan parasite trichomonas vaginalis in complex with mre-1-mre-2r DNA
- Reference
- Lou YC, Wei SY, Rajasekaran M, Chou CC, Hsu HM, Tai JH, Chen C (2009): "NMR structural analysis of DNA recognition by a novel Myb1 DNA-binding domain in the protozoan parasite Trichomonas vaginalis." Nucleic Acids Res. doi: 10.1093/nar/gkp097.
- Abstract
- The transcription regulator, tvMyb1, is the first Myb family protein identified in Trichomonas vaginalis. Using an electrophoretic mobility shift assay, we defined the amino-acid sequence from Lys(35) to Ser(141) (tvMyb1(35-141)) as the minimal DNA-binding domain, encompassing two Myb-like DNA-binding motifs (designated as R2 and R3 motifs) and an extension of 10 residues at the C-terminus. NMR solution structures of tvMyb1(35-141) show that both the R2 and R3 motifs adopt helix-turn-helix conformations while helix 6 in the R3 motif is longer than its counterpart in vertebrate Myb proteins. The extension of helix 6 was then shown to play an important role in protein stability as well as in DNA-binding activity. The structural basis for the tvMyb1(35-141)/DNA interaction was investigated using chemical shift perturbations, residual dipolar couplings, DNA specificity data and data-driven macromolecular docking by HADDOCK. Our data indicate that the orientation between R2 and R3 motifs dramatically changes upon binding to DNA so as to recognize the DNA major groove through a number of key contacts involving residues in helices 3 and 6. The tvMyb1(35-141)/DNA complex model furthers our understanding of DNA recognition by Myb proteins and this approach could be applied in determining the complex structures involving proteins with multiple domains.