Summary information and primary citation
- PDB-id
- 2kg1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- NMR
- Summary
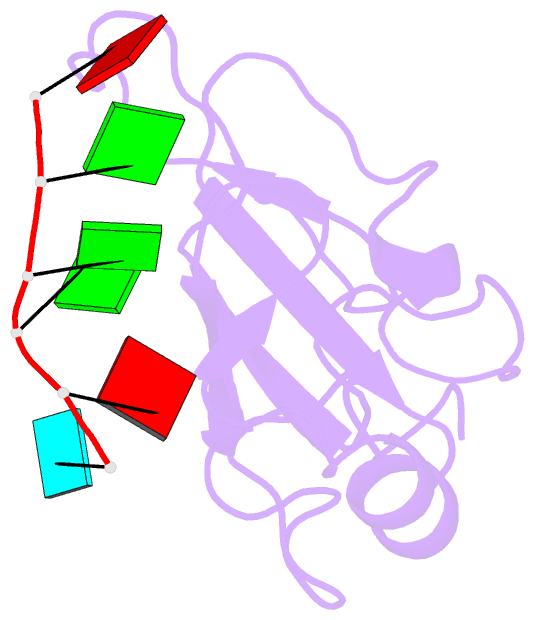
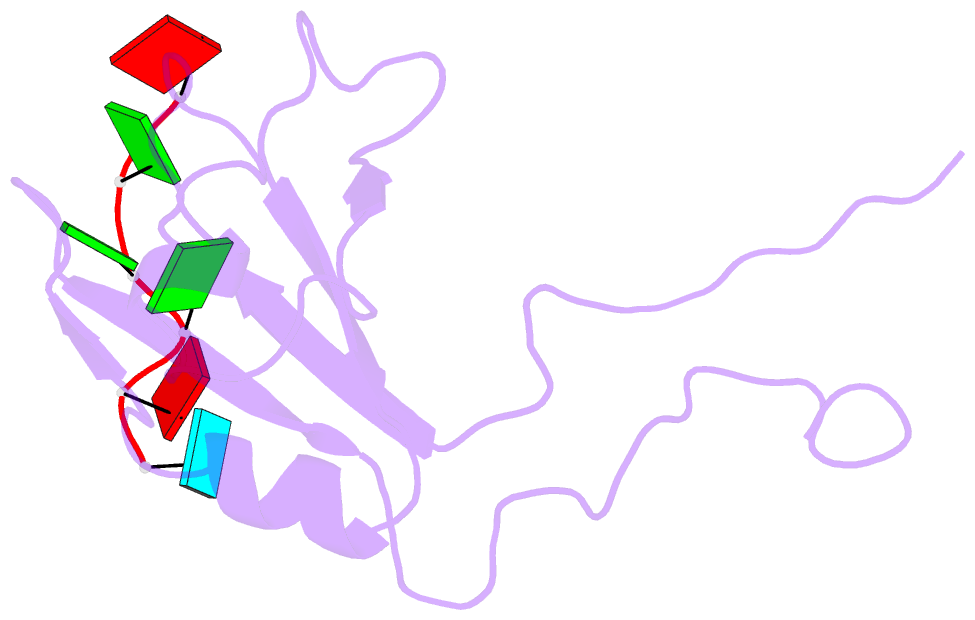
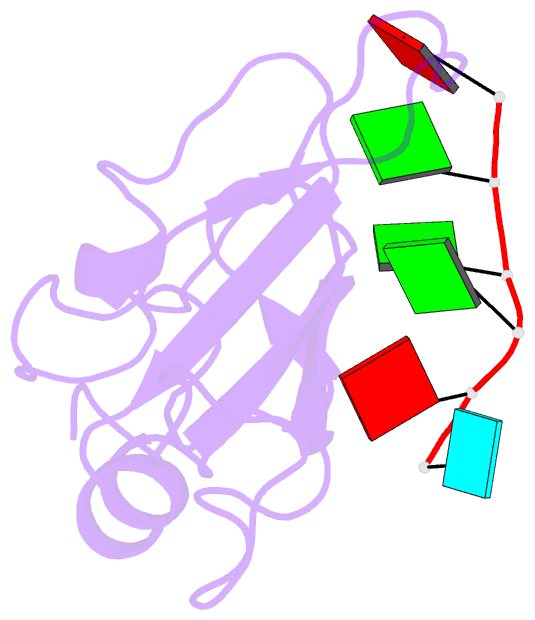
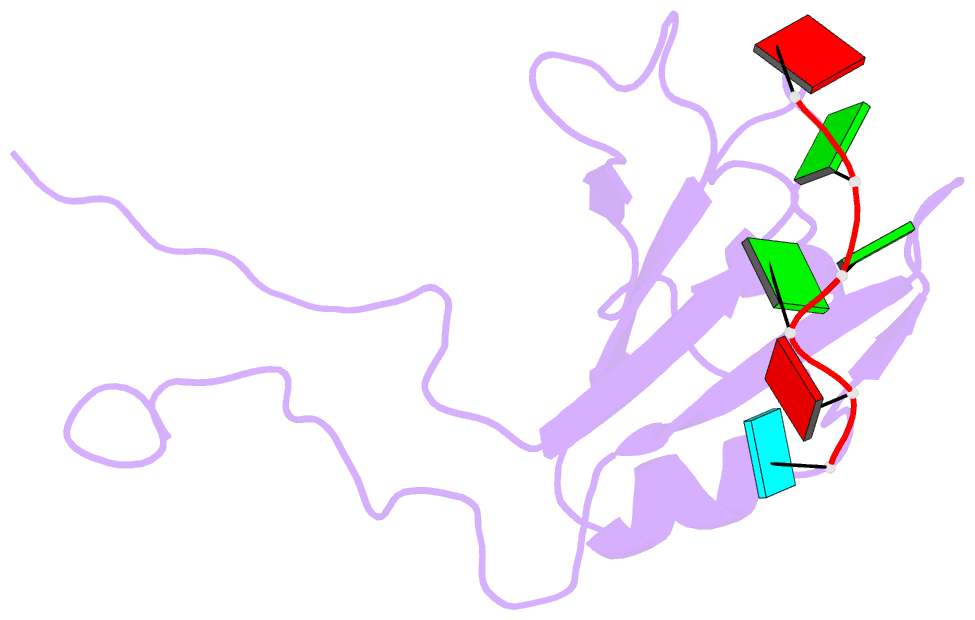
- Structure of the third qrrm domain of hnrnp f in complex with a agggau g-tract RNA
- Reference
- Dominguez C, Fisette JF, Chabot B, Allain FH (2010): "Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs." Nat.Struct.Mol.Biol., 17, 853-861. doi: 10.1038/nsmb.1814.
- Abstract
- The heterogeneous nuclear ribonucleoprotein (hnRNP) F is involved in the regulation of mRNA metabolism by specifically recognizing G-tract RNA sequences. We have determined the solution structures of the three quasi-RNA-recognition motifs (qRRMs) of hnRNP F in complex with G-tract RNA. These structures show that qRRMs bind RNA in a very unusual manner, with the G-tract 'encaged', making the qRRM a novel RNA binding domain. We defined a consensus signature sequence for qRRMs and identified other human qRRM-containing proteins that also specifically recognize G-tract RNAs. Our structures explain how qRRMs can sequester G-tracts, maintaining them in a single-stranded conformation. We also show that isolated qRRMs of hnRNP F are sufficient to regulate the alternative splicing of the Bcl-x pre-mRNA, suggesting that hnRNP F would act by remodeling RNA secondary and tertiary structures.