Summary information and primary citation
- PDB-id
- 2kmj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA-peptide
- Method
- NMR
- Summary
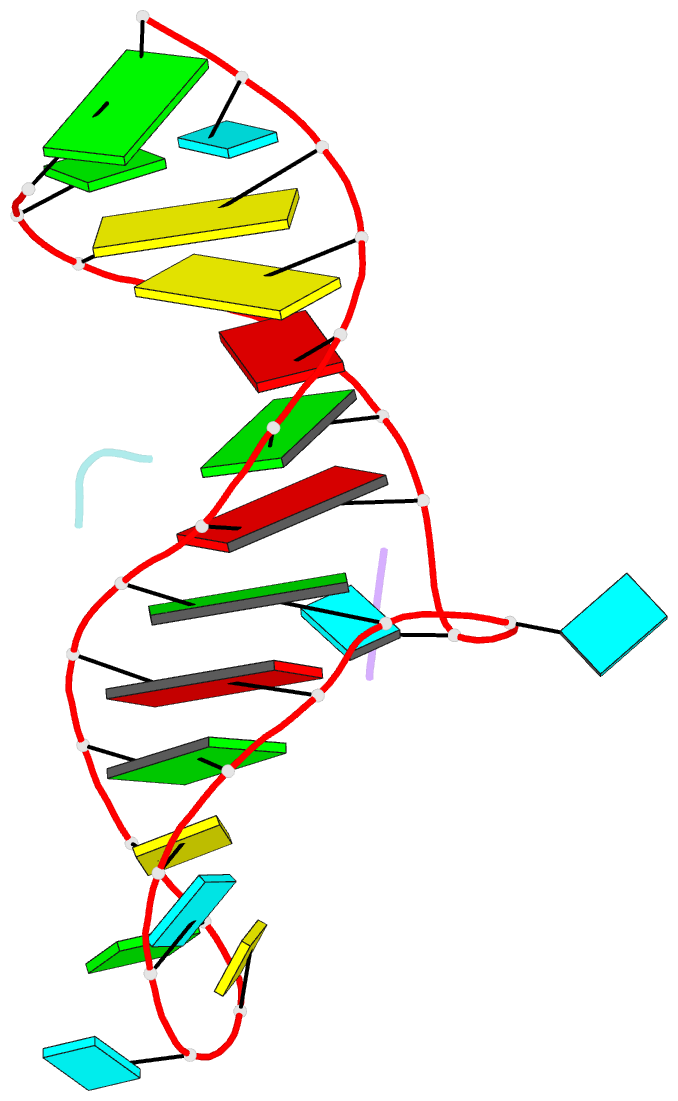
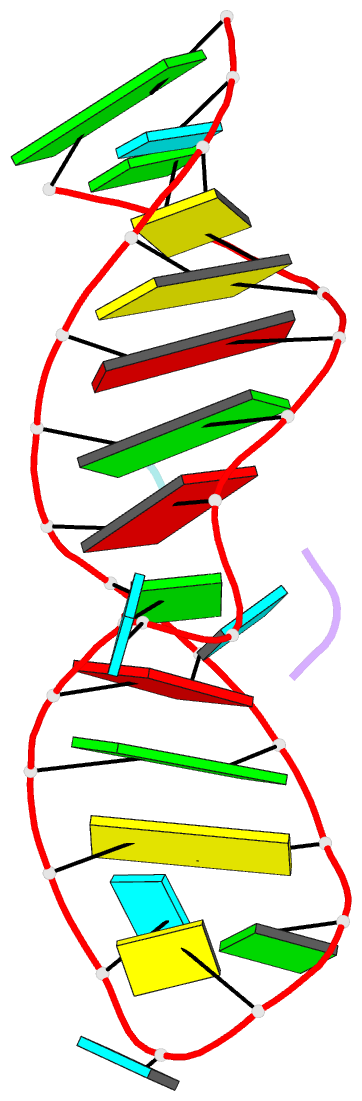
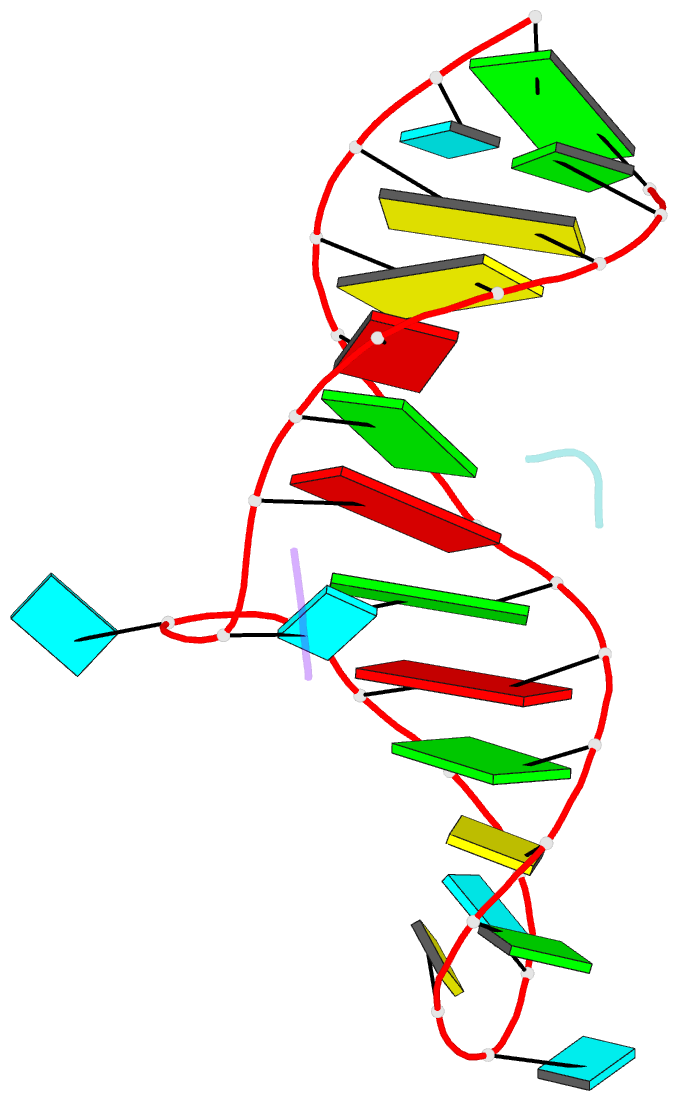
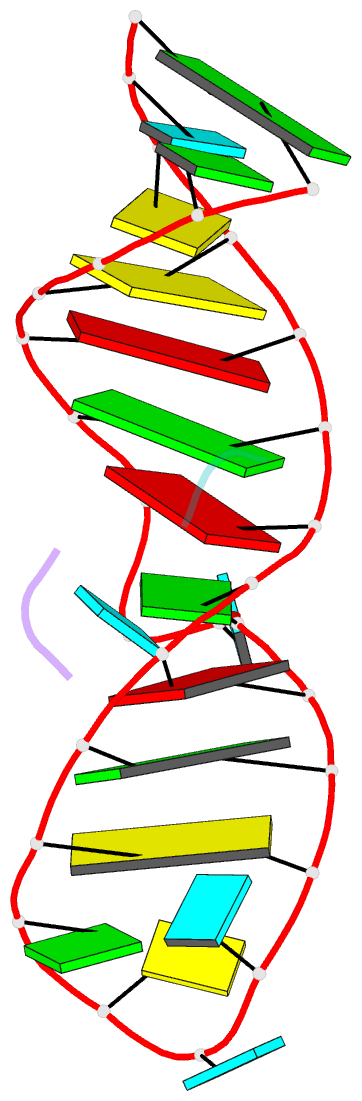
- High resolution NMR solution structure of a complex of hiv-2 tar RNA and a synthetic tripeptide in a 1:2 stoichiometry
- Reference
- Ferner J, Suhartono M, Breitung S, Jonker HRA, Hennig M, Wohnert J, Gobel M, Schwalbe H (2009): "Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries." Chembiochem, 10, 1490-1494. doi: 10.1002/cbic.200900220.
- Abstract
- Target TAR by NMR: Tripeptides containing arginines as terminal residues and non-natural amino acids as central residues are good leads for drug design to target the HIV trans-activation response element (TAR). The structural characterization of the RNA-ligand complex by NMR spectroscopy reveals two specific binding sites that are located at bulge residue U23 and around the pyrimidine-stretch U40-C41-U42 directly adjacent to the bulge.