Summary information and primary citation
- PDB-id
- 2ko0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- NMR
- Summary
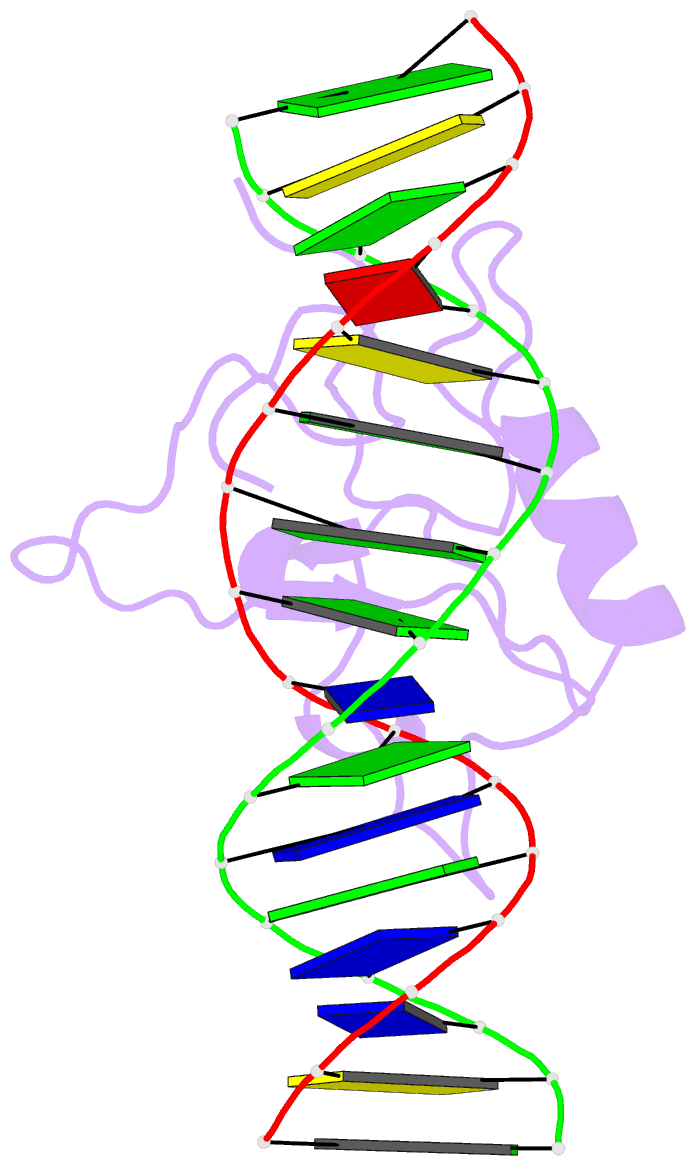
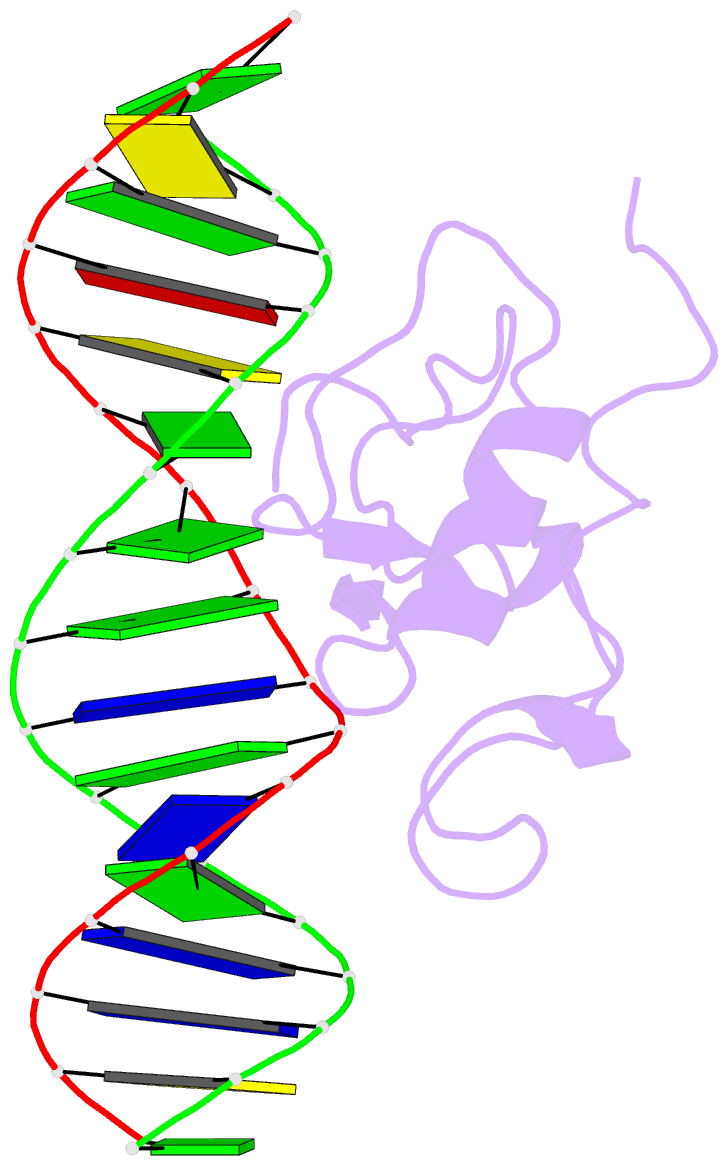
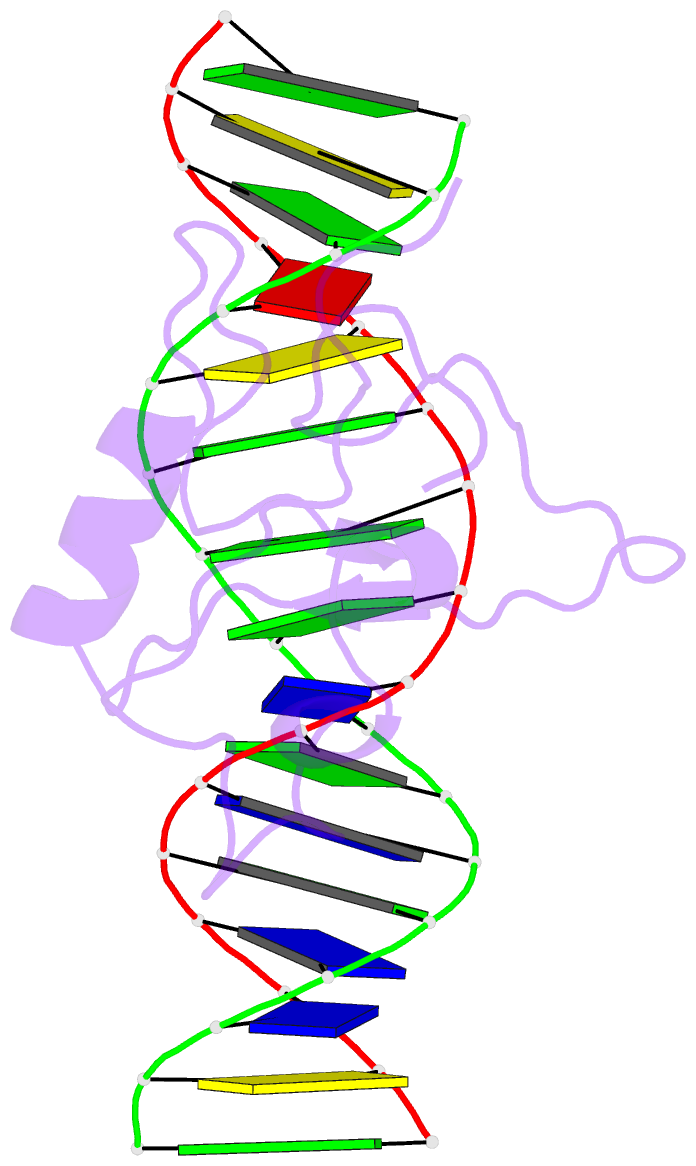
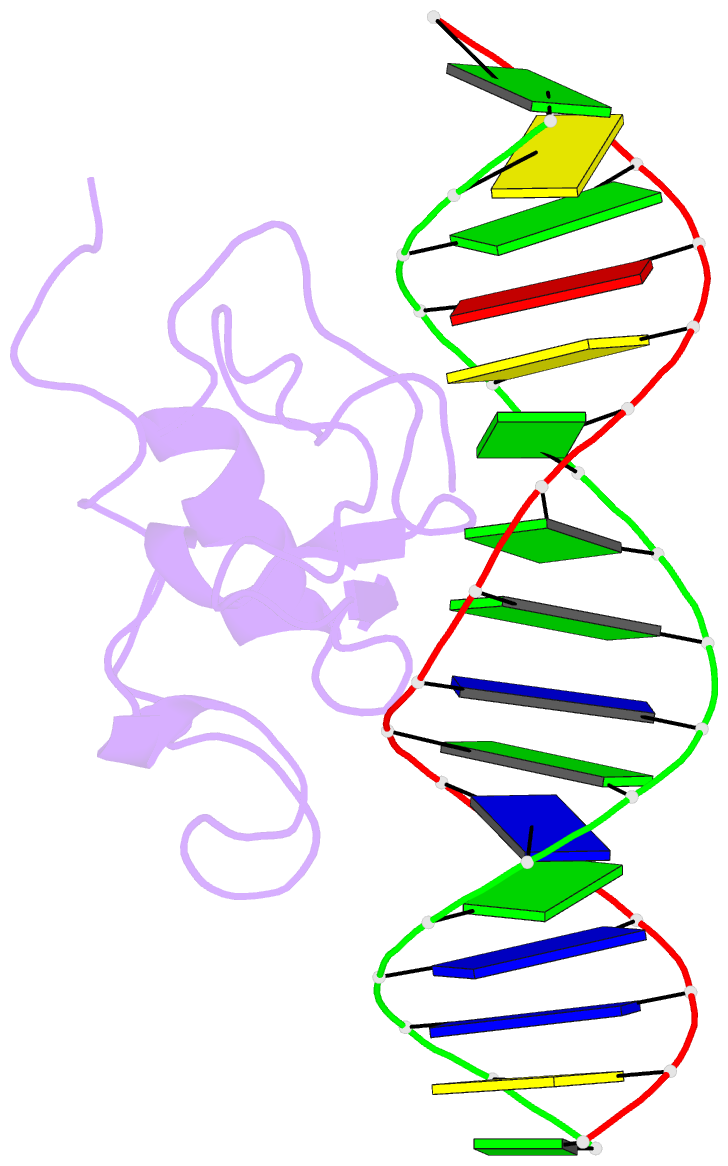
- Solution structure of the thap zinc finger of thap1 in complex with its DNA target
- Reference
- Campagne S, Saurel O, Gervais V, Milon A (2010): "Structural determinants of specific DNA-recognition by the THAP zinc finger." Nucleic Acids Res. doi: 10.1093/nar/gkq053.
- Abstract
- Human THAP1 is the prototype of a large family of cellular factors sharing an original THAP zinc-finger motif responsible for DNA binding. Human THAP1 regulates endothelial cell proliferation and G1/S cell-cycle progression, through modulation of pRb/E2F cell-cycle target genes including rrm1. Recently, mutations in THAP1 have been found to cause DYT6 primary torsion dystonia, a human neurological disease. We report here the first 3D structure of the complex formed by the DNA-binding domain of THAP1 and its specific DNA target (THABS) found within the rrm1 target gene. The THAP zinc finger uses its double-stranded beta-sheet to fill the DNA major groove and provides a unique combination of contacts from the beta-sheet, the N-terminal tail and surrounding loops toward the five invariant base pairs of the THABS sequence. Our studies reveal unprecedented insights into the specific DNA recognition mechanisms within this large family of proteins controlling cell proliferation, cell cycle and pluripotency.