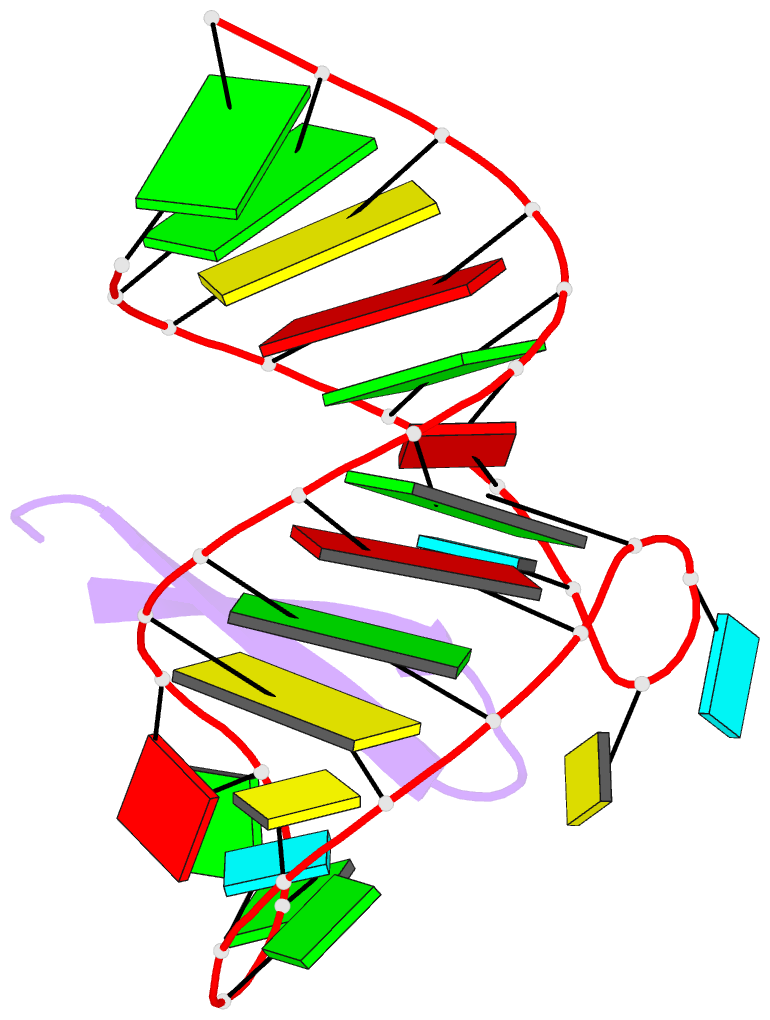
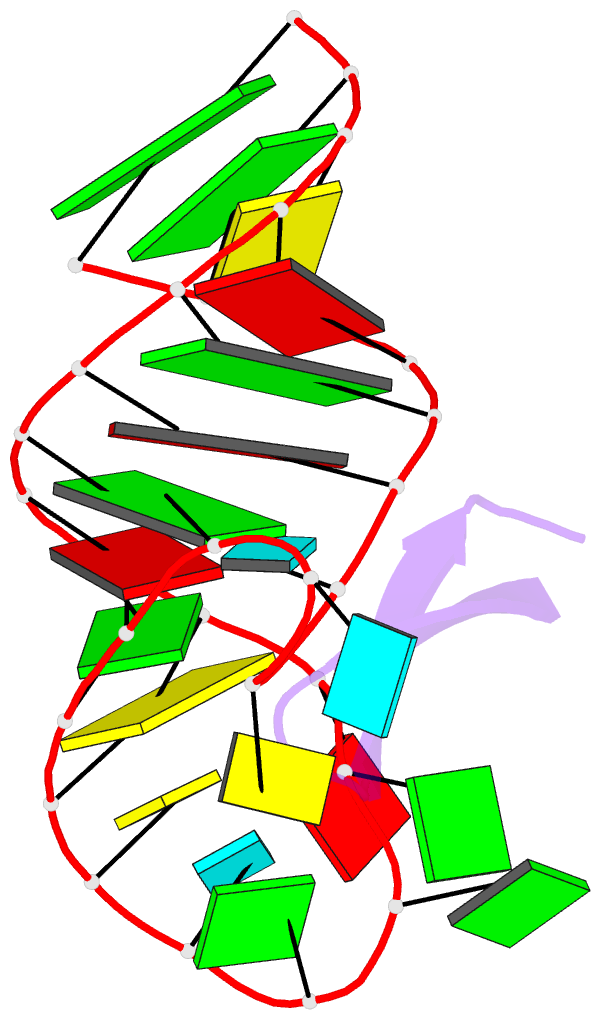
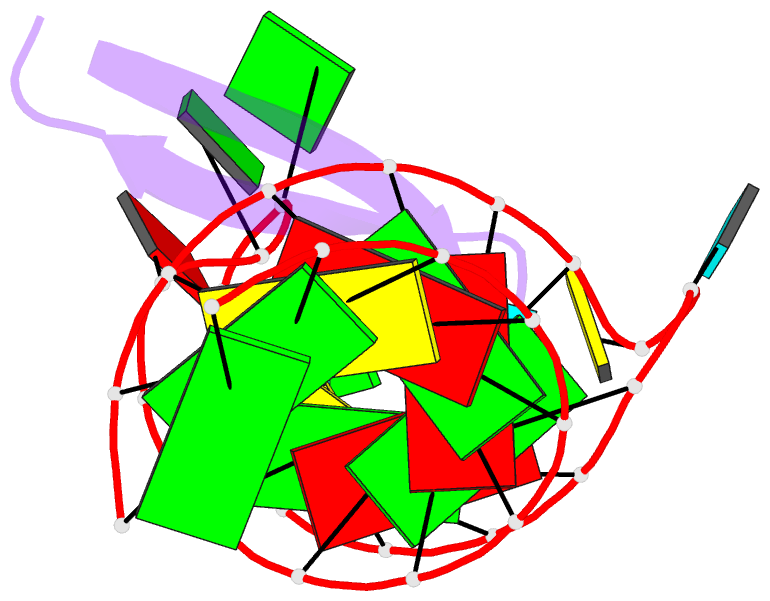
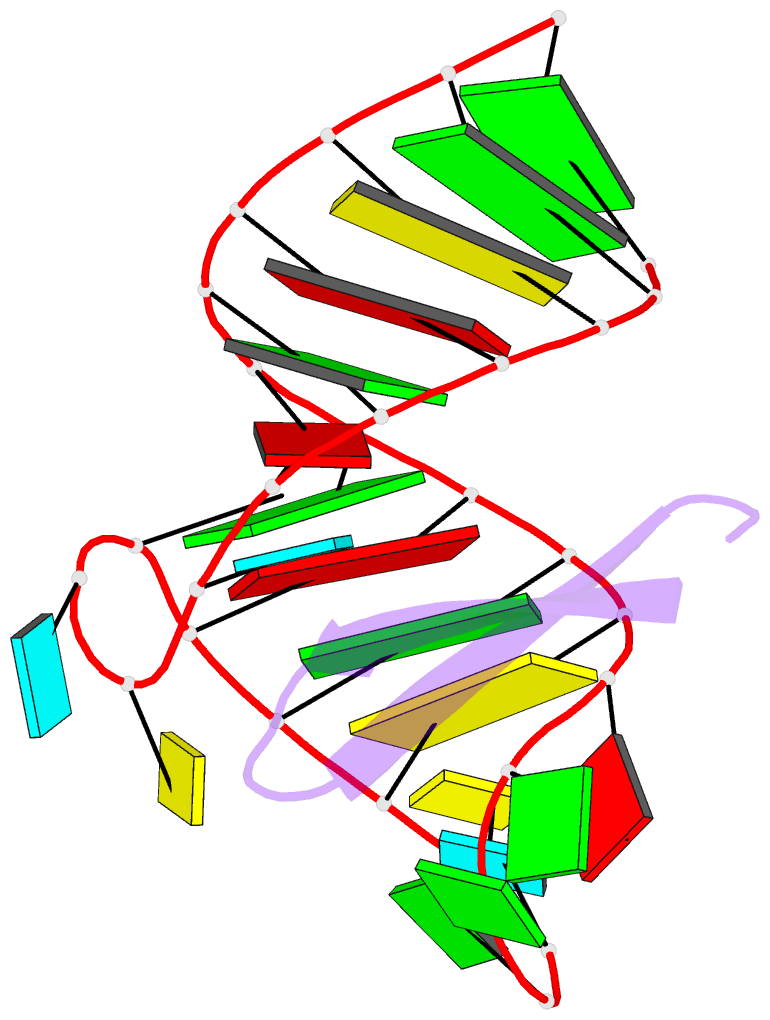
Summary information and primary citation
- PDB-id
- 2kx5; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- NMR
- Summary
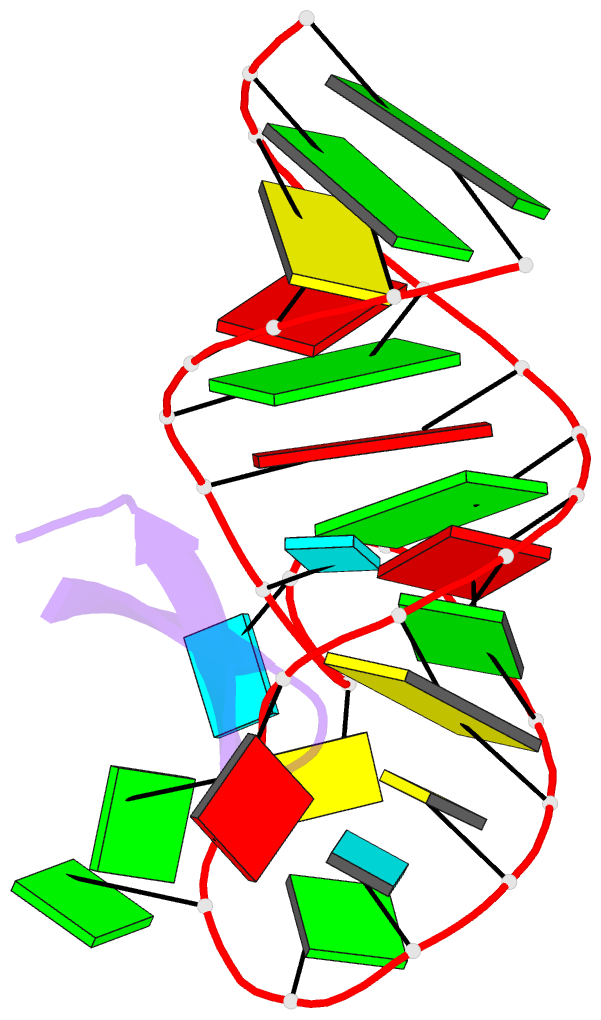
- Recognition of hiv tar RNA by peptide mimetic of tat protein
- Reference
- Davidson A, Patora-Komisarska K, Robinson JA, Varani G (2011): "Essential structural requirements for specific recognition of HIV TAR RNA by peptide mimetics of Tat protein." Nucleic Acids Res., 39, 248-256. doi: 10.1093/nar/gkq713.
- Abstract
- The pharmacological disruption of the interaction between the HIV Tat protein and its cognate transactivation response RNA (TAR) would generate novel anti-viral drugs with a low susceptibility to drug resistance, but efforts to discover ligands with sufficient potency to warrant pharmaceutical development have been unsuccessful. We have previously described a family of structurally constrained β-hairpin peptides that potently inhibits viral growth in HIV-infected cells. The nuclear magnetic resonance (NMR) structure of an inhibitory complex revealed that the peptide makes intimate contacts with the 3-nt bulge and the upper helix of the RNA hairpin, but that a single residue contacts the apical loop where recruitment of the essential cellular co-factor cyclin T₁ occurs. Attempting to extend the peptide to form more interactions with the RNA loop, we examined a library of longer peptides and achieved > 6-fold improvement in affinity. The structure of TAR bound to one of the extended peptides reveals that the peptide slides down the major groove of the RNA, relative to our design, in order to maintain critical interactions with TAR. These conserved contacts involve three amino acid side chains and identify critical interaction points required for potent and specific binding to TAR RNA. They constitute a template of essential interactions required for inhibition of this RNA.