Summary information and primary citation
- PDB-id
- 2l4l; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-DNA
- Method
- NMR
- Summary
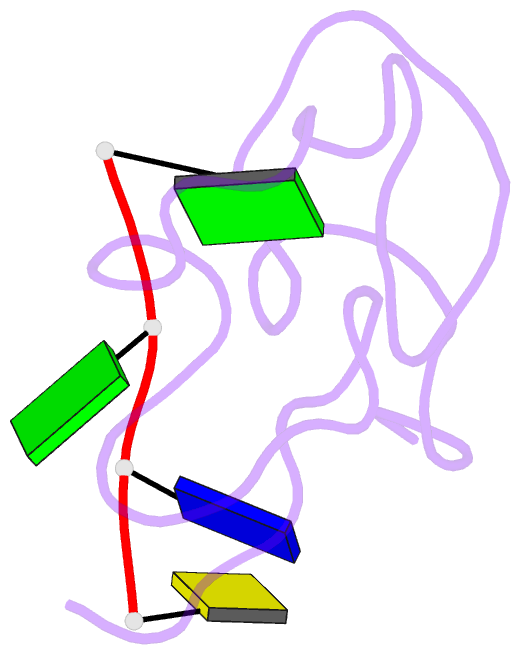
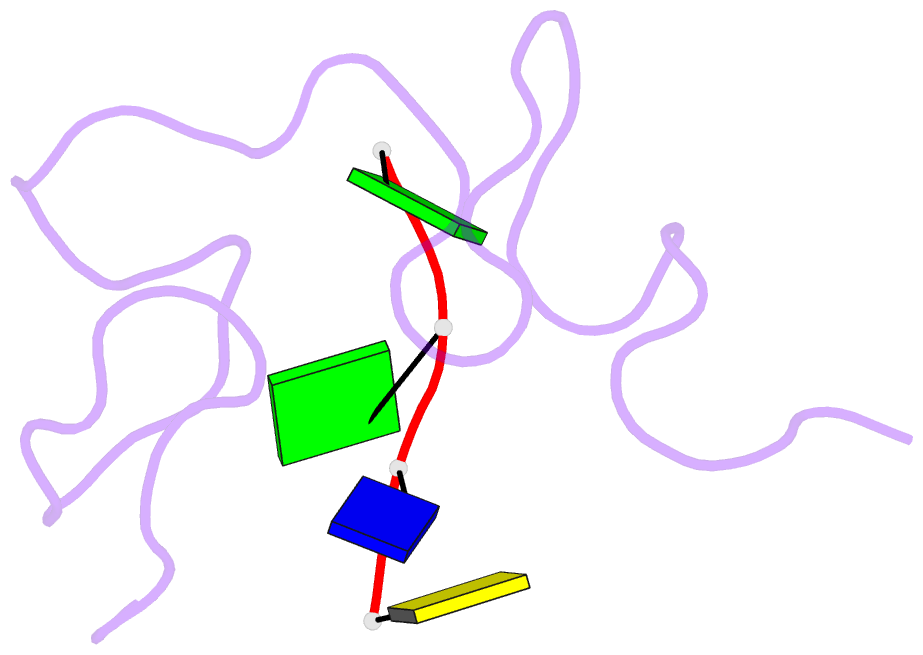
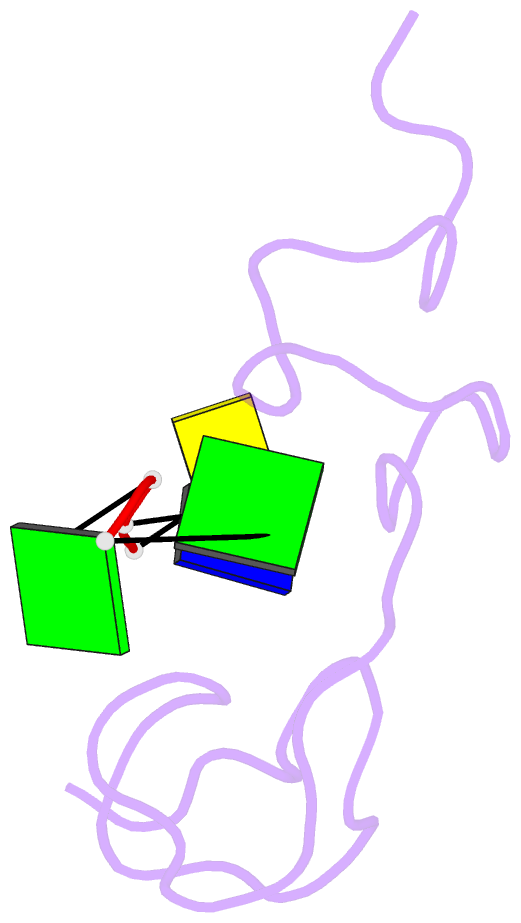
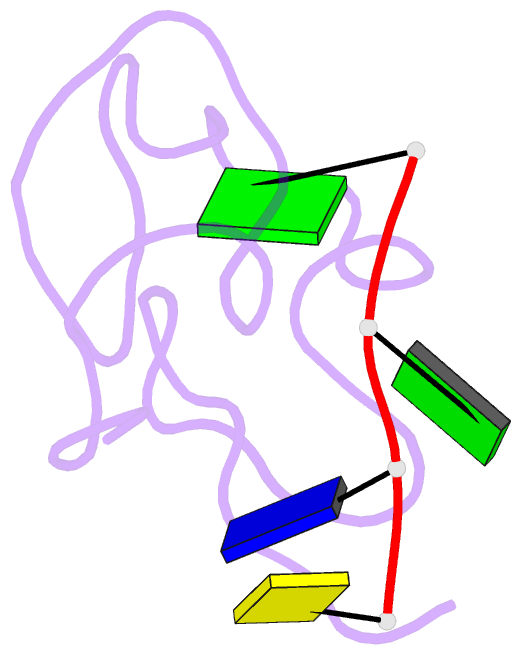
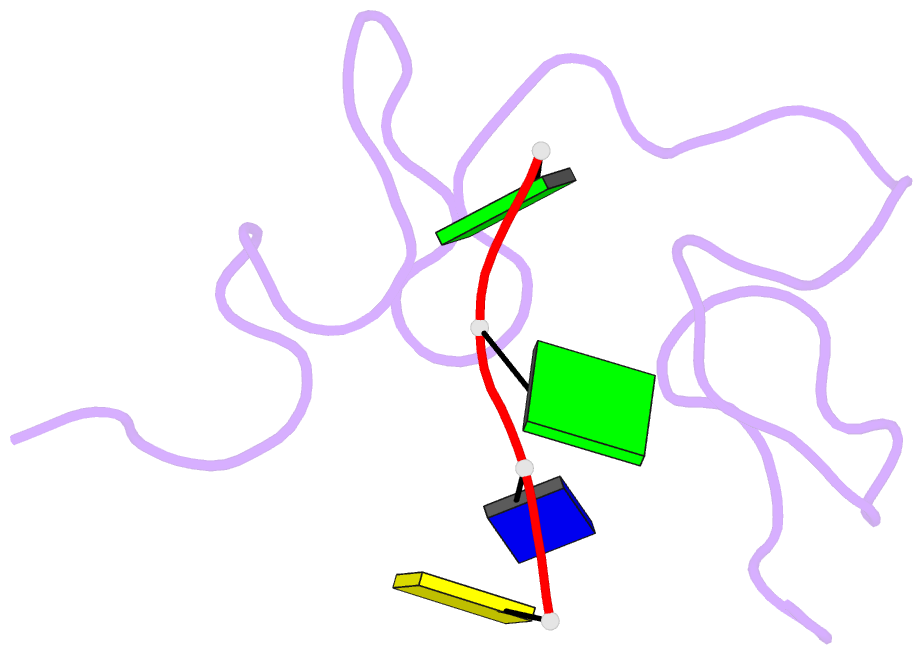
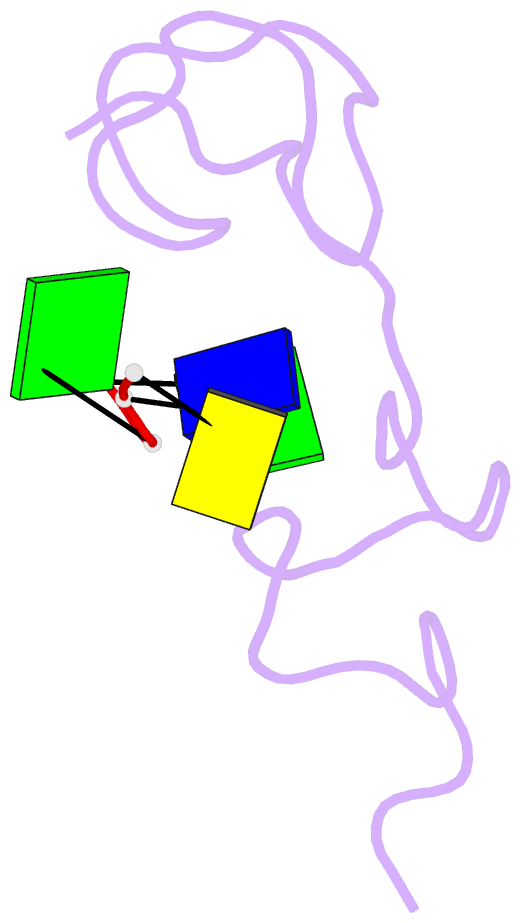
- Structural insights into the ctar DNA recognition by the hiv-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of nc
- Reference
- Bazzi A, Zargarian L, Chaminade F, Boudier C, De Rocquigny H, Rene B, Mely Y, Fosse P, Mauffret O (2011): "Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC." Nucleic Acids Res., 39, 3903-3916. doi: 10.1093/nar/gkq1290.
- Abstract
- An essential step of the reverse transcription of the HIV-1 genome is the first strand transfer that requires the annealing of the TAR RNA hairpin to the cTAR DNA hairpin. HIV-1 nucleocapsid protein (NC) plays a crucial role by facilitating annealing of the complementary hairpins. Using nuclear magnetic resonance and gel retardation assays, we investigated the interaction between NC and the top half of the cTAR DNA (mini-cTAR). We show that NC(11-55) binds the TGG sequence in the lower stem that is destabilized by the adjacent internal loop. The 5' thymine interacts with residues of the N-terminal zinc knuckle and the 3' guanine is inserted in the hydrophobic plateau of the C-terminal zinc knuckle. The TGG sequence is preferred relative to the apical and internal loops containing unpaired guanines. Investigation of the DNA-protein contacts shows the major role of hydrophobic interactions involving nucleobases and deoxyribose sugars. A similar network of hydrophobic contacts is observed in the published NC:DNA complexes, whereas NC contacts ribose differently in NC:RNA complexes. We propose that the binding polarity of NC is related to these contacts that could be responsible for the preferential binding to single-stranded nucleic acids.