Summary information and primary citation
- PDB-id
- 2lup; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- NMR
- Summary
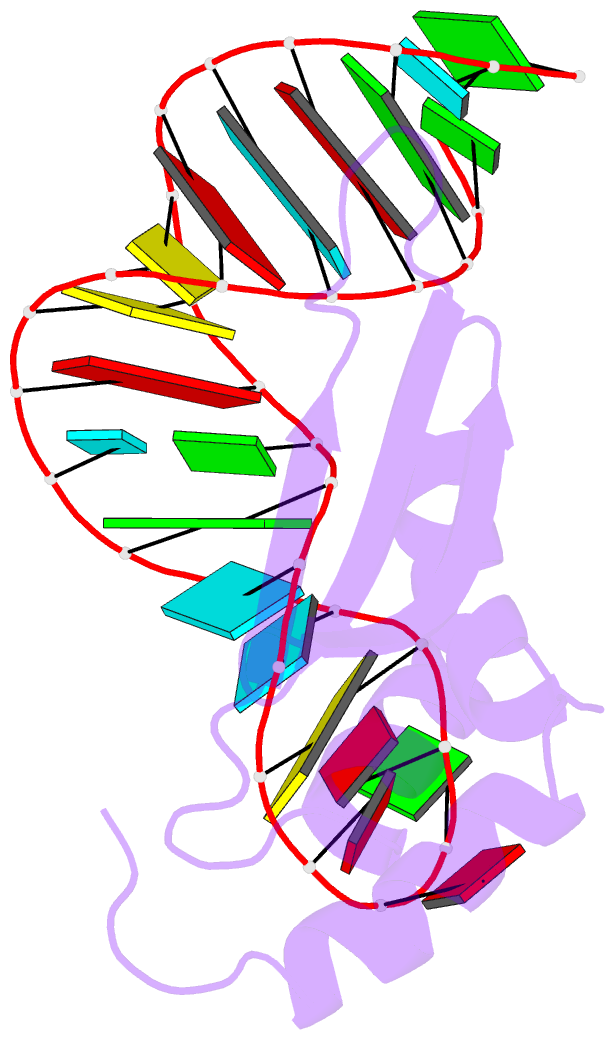
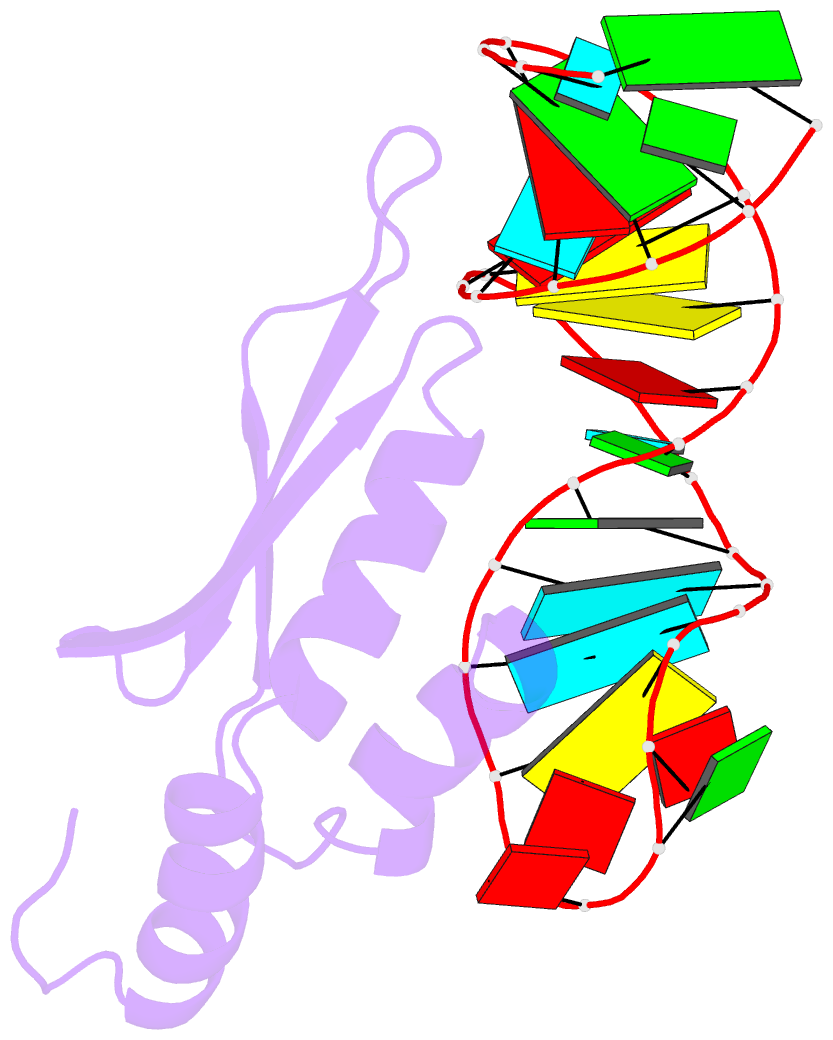
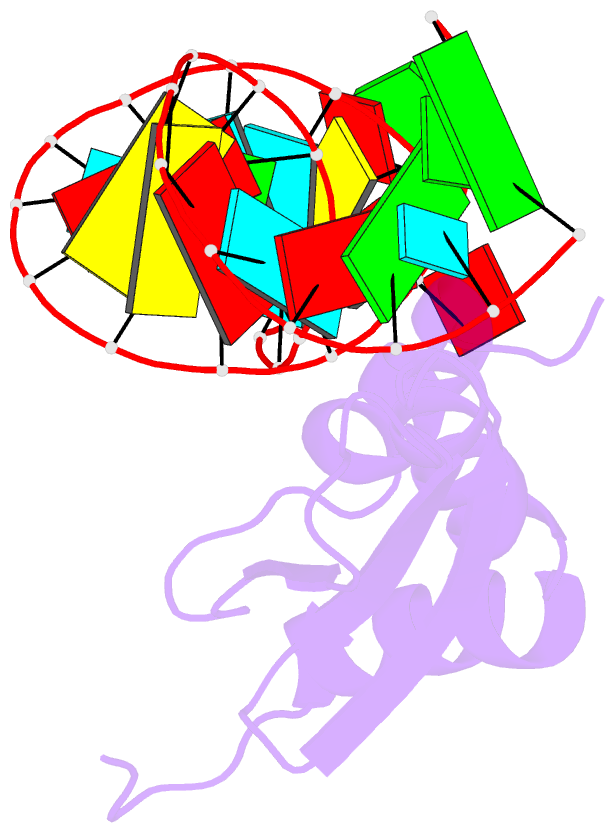
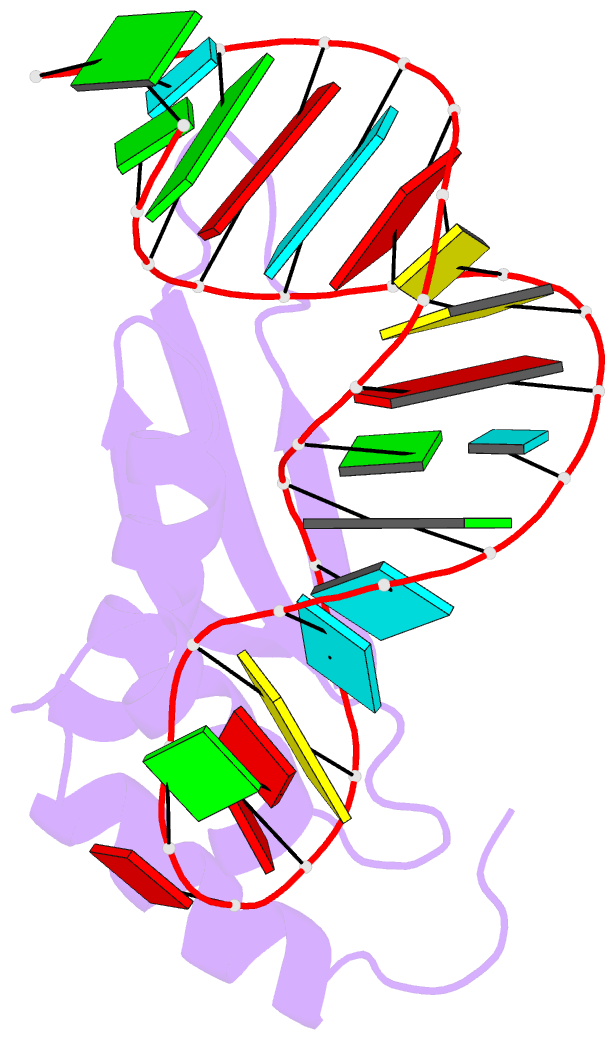
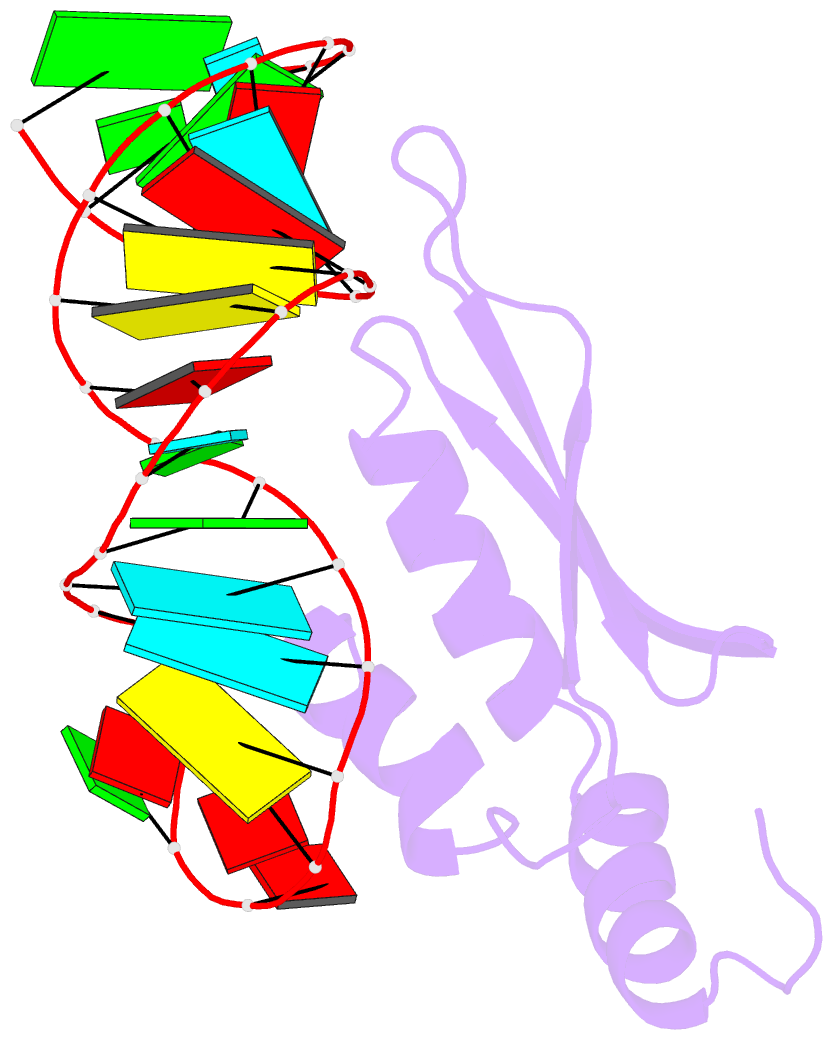
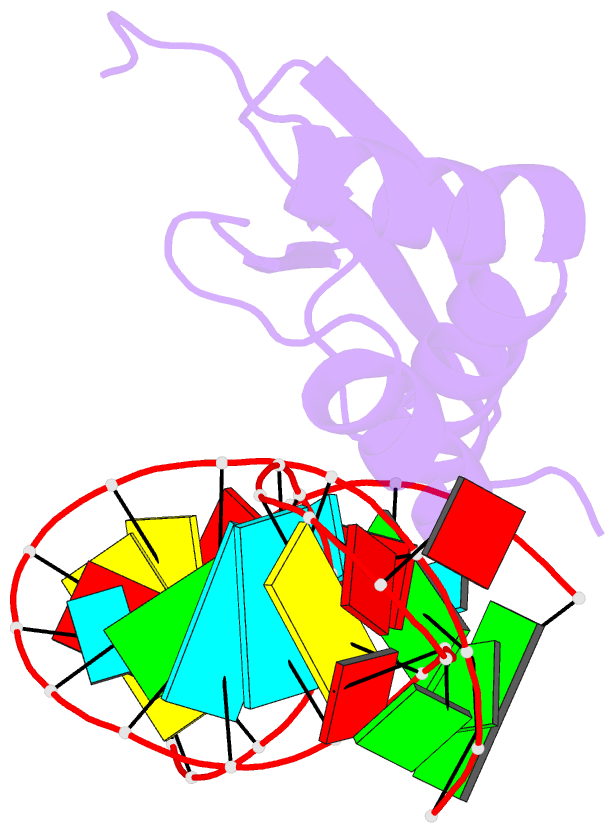
- Rdc refined solution structure of double-stranded RNA binding domain of s. cerevisiae rnase iii (rnt1p) in complex with the terminal RNA hairpin of snr47 precursor
- Reference
- Wang Z, Hartman E, Roy K, Chanfreau G, Feigon J (2011): "Structure of a yeast RNase III dsRBD complex with a noncanonical RNA substrate provides new insights into binding specificity of dsRBDs." Structure, 19, 999-1010. doi: 10.1016/j.str.2011.03.022.
- Abstract
- dsRBDs often bind dsRNAs with some specificity, yet the basis for this is poorly understood. Rnt1p, the major RNase III in Saccharomyces cerevisiae, cleaves RNA substrates containing hairpins capped by A/uGNN tetraloops, using its dsRBD to recognize a conserved tetraloop fold. However, the identification of a Rnt1p substrate with an AAGU tetraloop raised the question of whether Rnt1p binds to this noncanonical substrate differently than to A/uGNN tetraloops. The solution structure of Rnt1p dsRBD bound to an AAGU-capped hairpin reveals that the tetraloop undergoes a structural rearrangement upon binding to Rnt1p dsRBD to adopt a backbone conformation that is essentially the same as the AGAA tetraloop, and indicates that a conserved recognition mode is used for all Rnt1p substrates. Comparison of free and RNA-bound Rnt1p dsRBD reveals that tetraloop-specific binding requires a conformational change in helix α1. Our findings provide a unified model of binding site selection by this dsRBD.