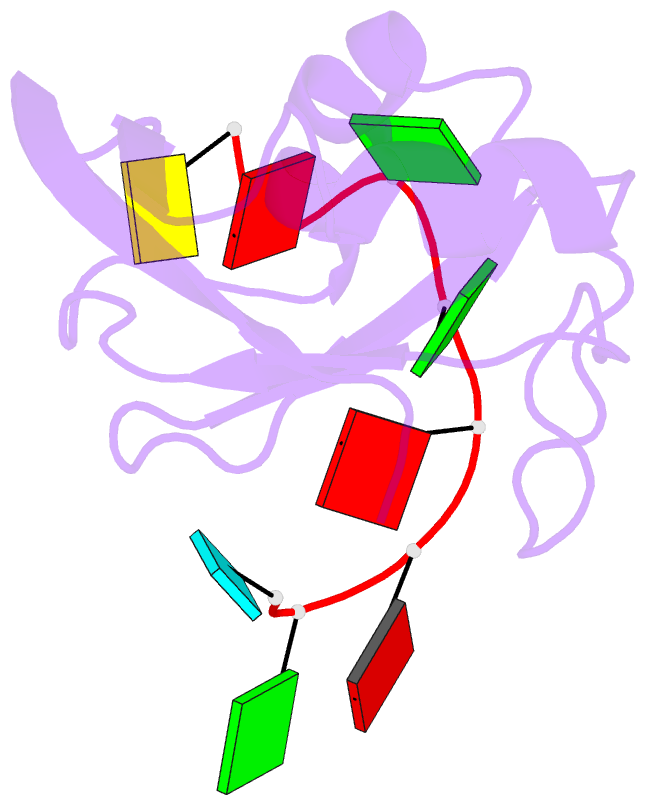
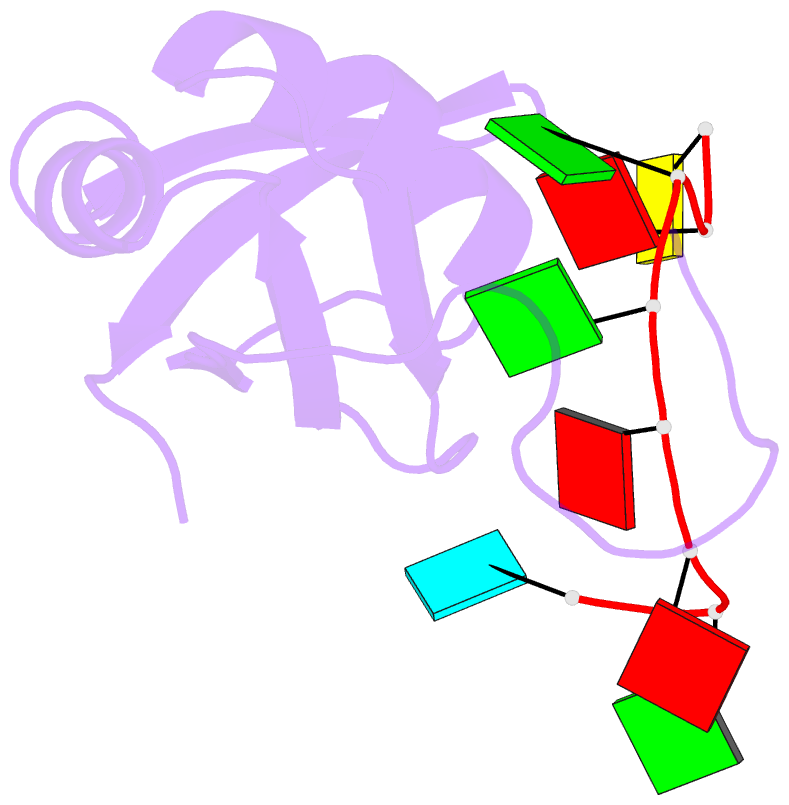
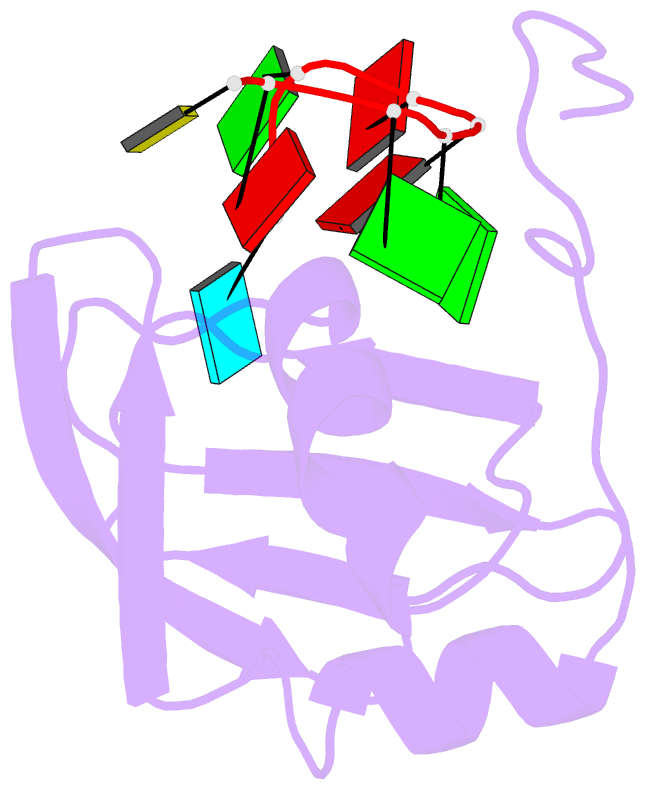
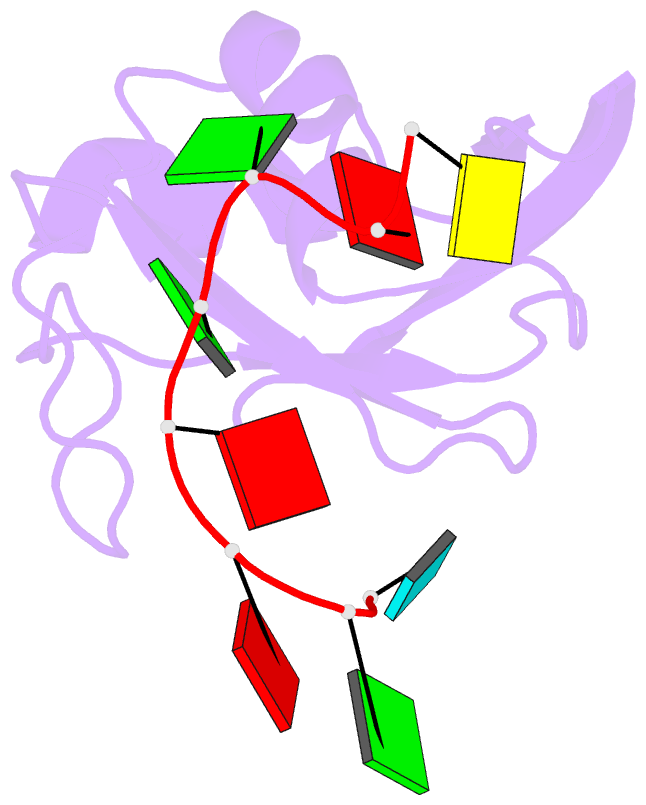
Summary information and primary citation
- PDB-id
- 2m8d; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- NMR
- Summary
- Structure of srsf1 rrm2 in complex with the RNA 5'-ugaaggac-3'
- Reference
- Clery A, Sinha R, Anczukow O, Corrionero A, Moursy A, Daubner GM, Valcarcel J, Krainer AR, Allain FH (2013): "Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition." Proc.Natl.Acad.Sci.USA, 110, E2802-E2811. doi: 10.1073/pnas.1303445110.
- Abstract
- Serine/arginine (SR) proteins, one of the major families of alternative-splicing regulators in Eukarya, have two types of RNA-recognition motifs (RRMs): a canonical RRM and a pseudo-RRM. Although pseudo-RRMs are crucial for activity of SR proteins, their mode of action was unknown. By solving the structure of the human SRSF1 pseudo-RRM bound to RNA, we discovered a very unusual and sequence-specific RNA-binding mode that is centered on one α-helix and does not involve the β-sheet surface, which typically mediates RNA binding by RRMs. Remarkably, this mode of binding is conserved in all pseudo-RRMs tested. Furthermore, the isolated pseudo-RRM is sufficient to regulate splicing of about half of the SRSF1 target genes tested, and the bound α-helix is a pivotal element for this function. Our results strongly suggest that SR proteins with a pseudo-RRM frequently regulate splicing by competing with, rather than recruiting, spliceosome components, using solely this unusual RRM.