Summary information and primary citation
- PDB-id
- 2mb0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- splicing-RNA
- Method
- NMR
- Summary
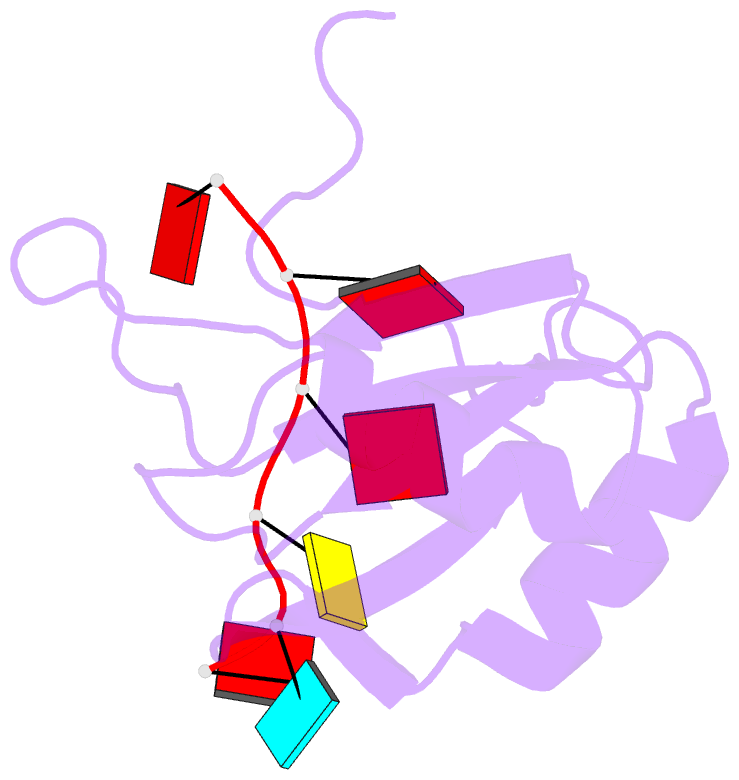
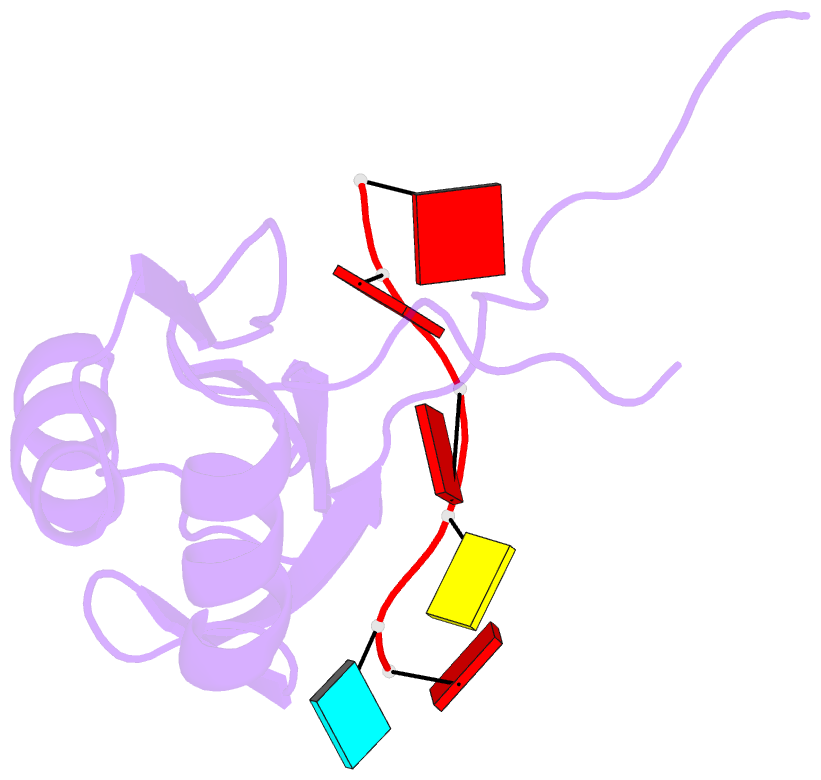
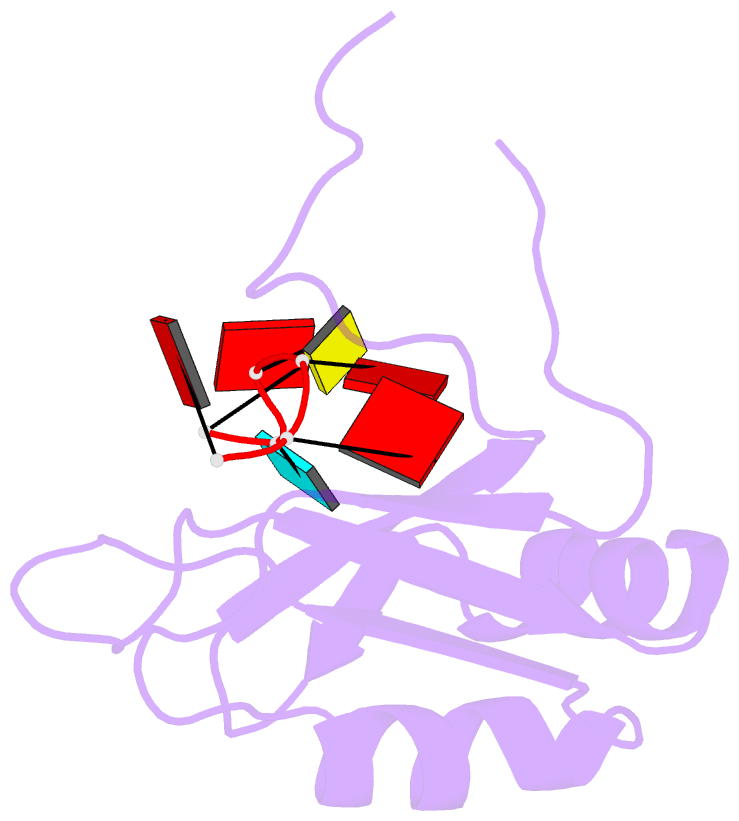
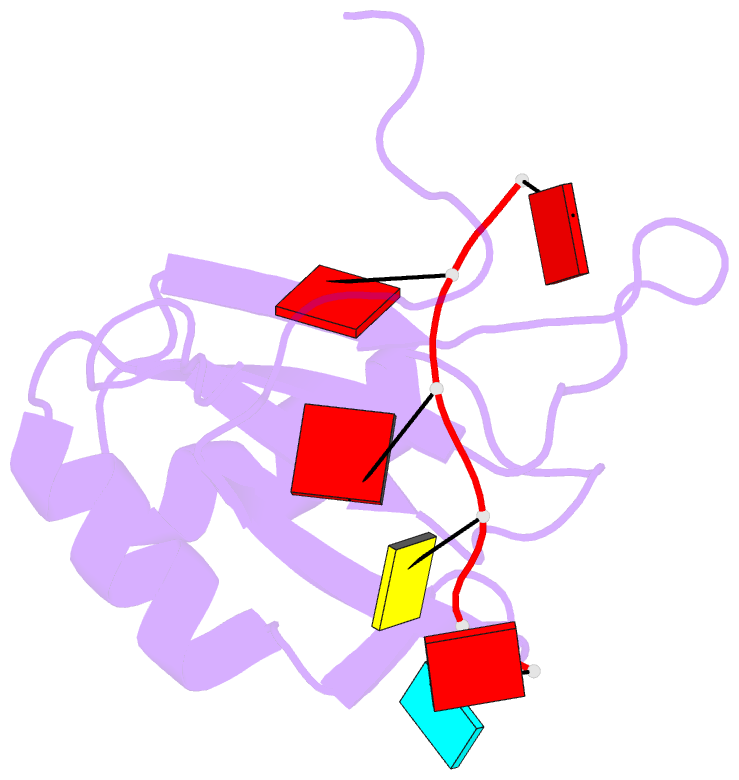
- Solution structure of hnrnp g rrm in complex with the RNA 5'-aucaaa-3'
- Reference
- Moursy A, Allain FH, Clery A (2014): "Characterization of the RNA recognition mode of hnRNP G extends its role in SMN2 splicing regulation." Nucleic Acids Res., 42, 6659-6672. doi: 10.1093/nar/gku244.
- Abstract
- Regulation of SMN2 exon 7 splicing is crucial for the production of active SMN protein and the survival of Spinal Muscular Atrophy (SMA) patients. One of the most efficient activators of exon 7 inclusion is hnRNP G, which is recruited to the exon by Tra2-β1. We report that in addition to the C-terminal region of hnRNP G, the RNA Recognition Motif (RRM) and the middle part of the protein containing the Arg-Gly-Gly (RGG) box are important for this function. To better understand the mode of action of hnRNP G in this context we determined the structure of its RRM bound to an SMN2 derived RNA. The RRM interacts with a 5'-AAN-3' motif and specifically recognizes the two consecutive adenines. By testing the effect of mutations in hnRNP G RRM and in its putative binding sites on the splicing of SMN2 exon 7, we show that it specifically binds to exon 7. This interaction is required for hnRNP G splicing activity and we propose its recruitment to a polyA tract located upstream of the Tra2-β1 binding site. Finally, our data suggest that hnRNP G plays a major role in the recruitment of the Tra2-β1/hnRNP G/SRSF9 trimeric complex to SMN2 exon 7.