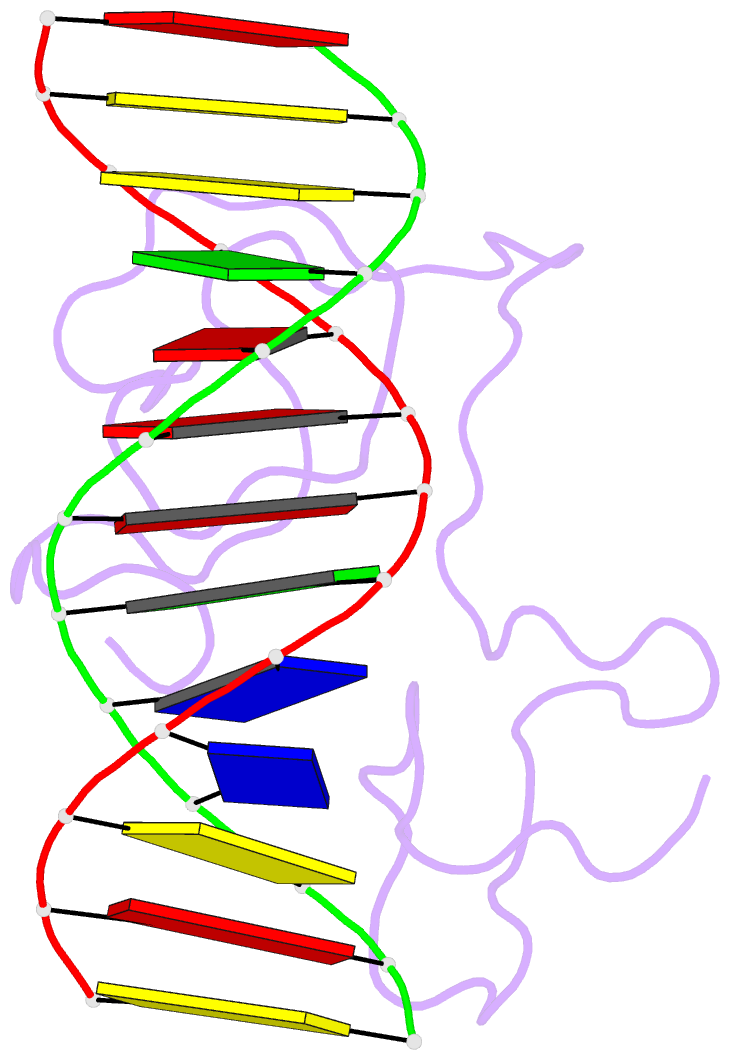
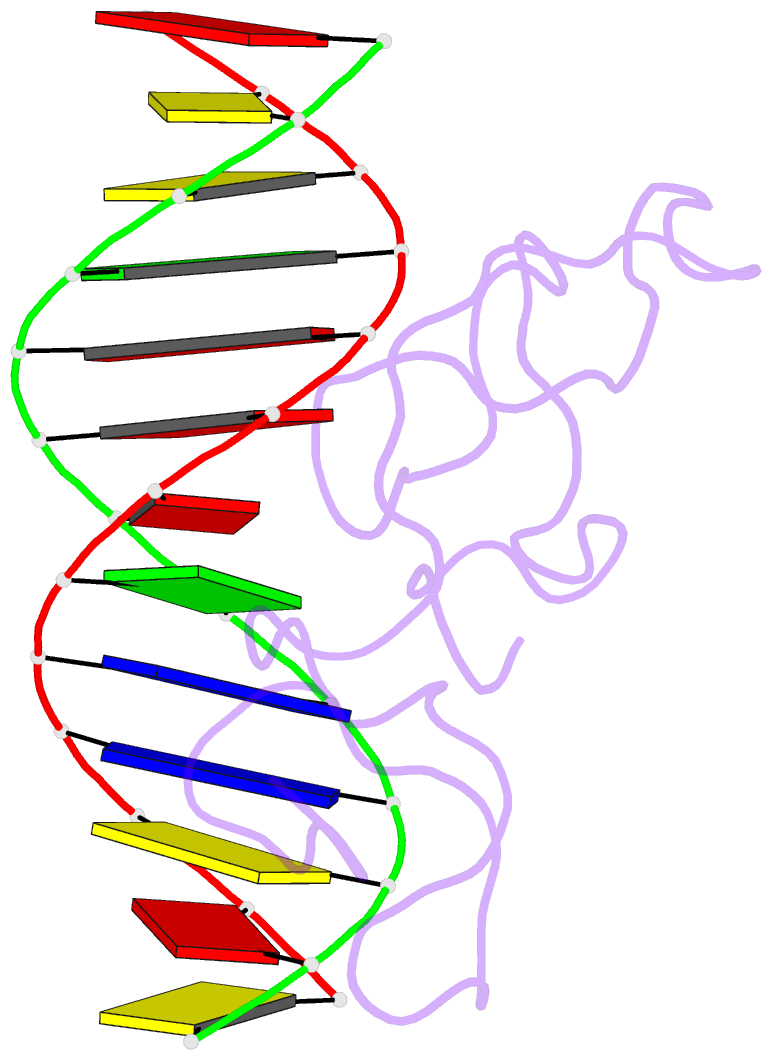
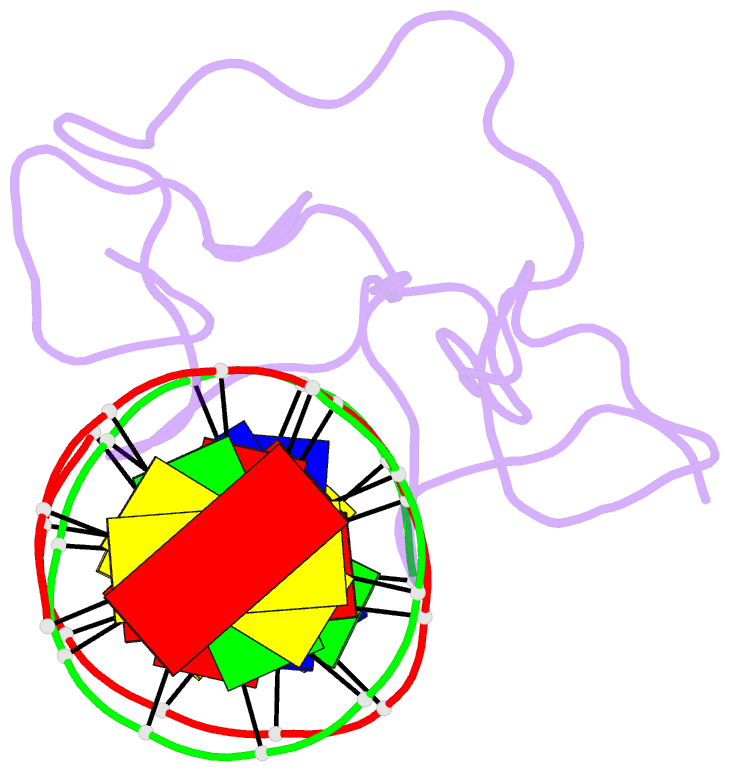
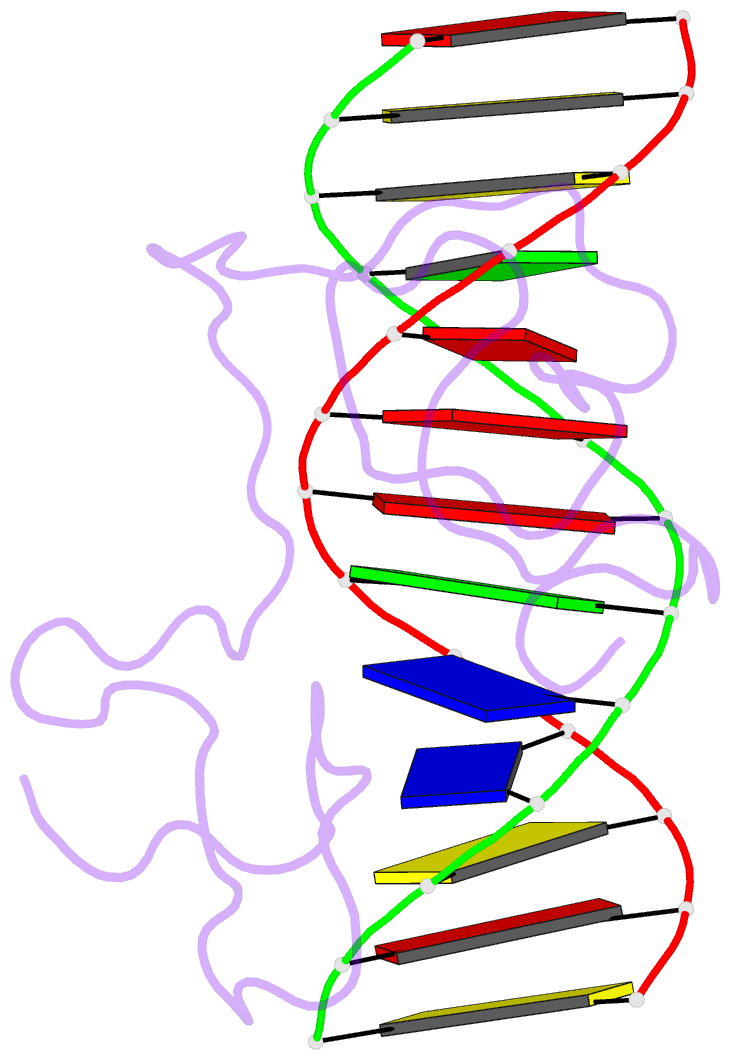
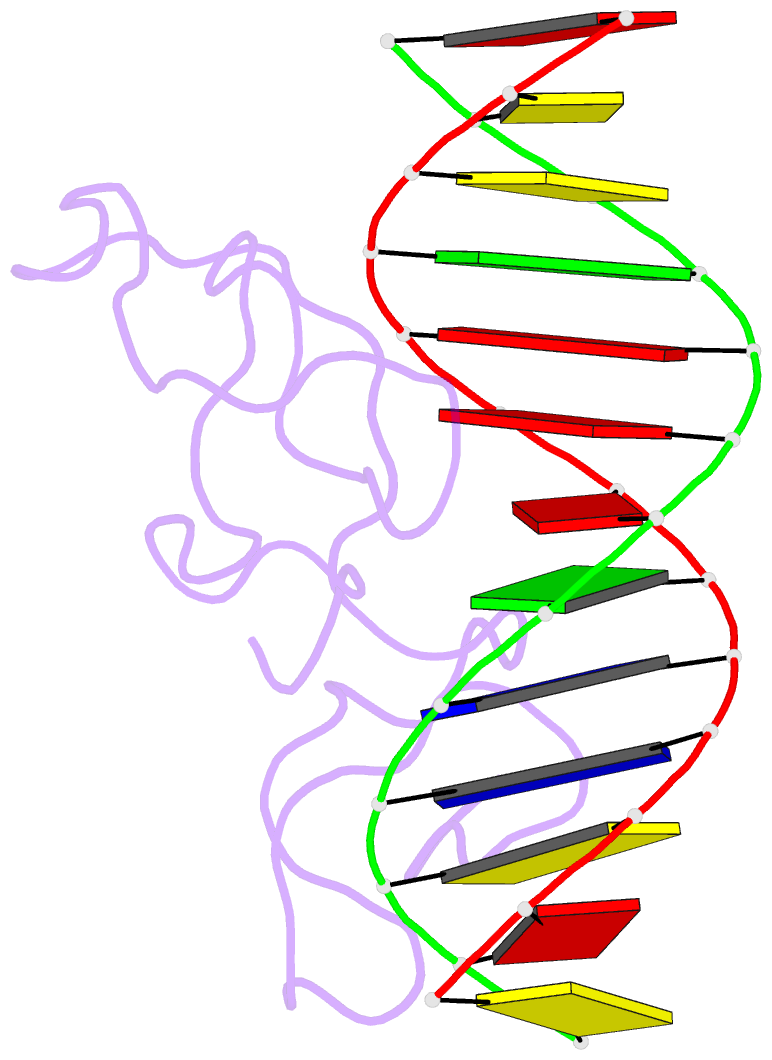
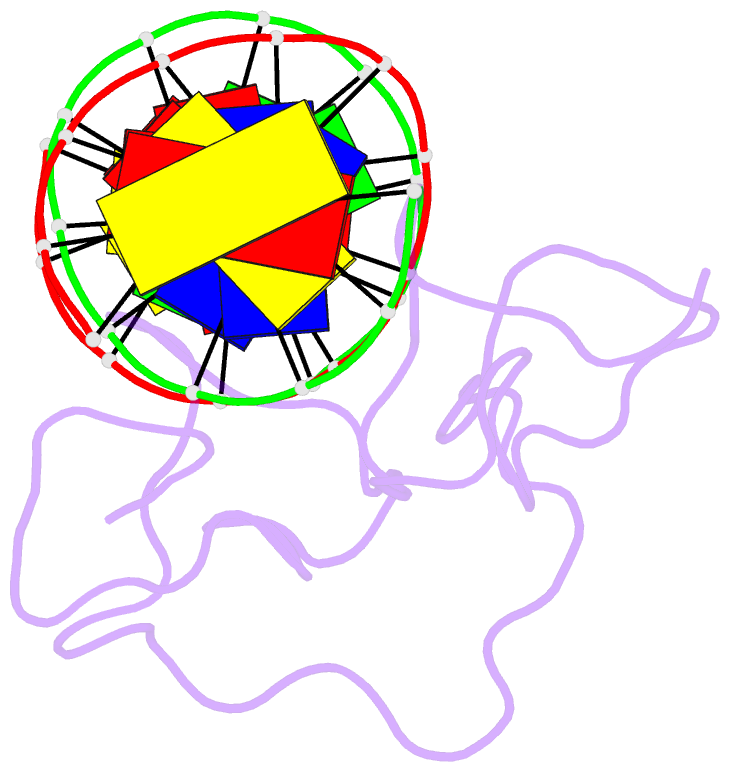
Summary information and primary citation
- PDB-id
- 2mf8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- metal binding protein-DNA
- Method
- NMR
- Summary
- Haddock model of myt1 f4f5 - DNA complex
- Reference
- Gamsjaeger R, O'Connell MR, Cubeddu L, Shepherd NE, Lowry JA, Kwan AH, Vandevenne M, Swanton MK, Matthews JM, Mackay JP (2013): "A structural analysis of DNA binding by myelin transcription factor 1 double zinc fingers." J.Biol.Chem., 288, 35180-35191. doi: 10.1074/jbc.M113.482075.
- Abstract
- Myelin transcription factor 1 (MyT1/NZF2), a member of the neural zinc-finger (NZF) protein family, is a transcription factor that plays a central role in the developing central nervous system. It has also recently been shown that, in combination with two other transcription factors, the highly similar paralog MyT1L is able to direct the differentiation of murine and human stem cells into functional neurons. MyT1 contains seven zinc fingers (ZFs) that are highly conserved throughout the protein and throughout the NZF family. We recently presented a model for the interaction of the fifth ZF of MyT1 with a DNA sequence derived from the promoter of the retinoic acid receptor (RARE) gene. Here, we have used NMR spectroscopy, in combination with surface plasmon resonance and data-driven molecular docking, to delineate the mechanism of DNA binding for double ZF polypeptides derived from MyT1. Our data indicate that a two-ZF unit interacts with the major groove of the entire RARE motif and that both fingers bind in an identical manner and with overall two-fold rotational symmetry, consistent with the palindromic nature of the target DNA. Several key residues located in one of the irregular loops of the ZFs are utilized to achieve specific binding. Analysis of the human and mouse genomes based on our structural data reveals three putative MyT1 target genes involved in neuronal development.