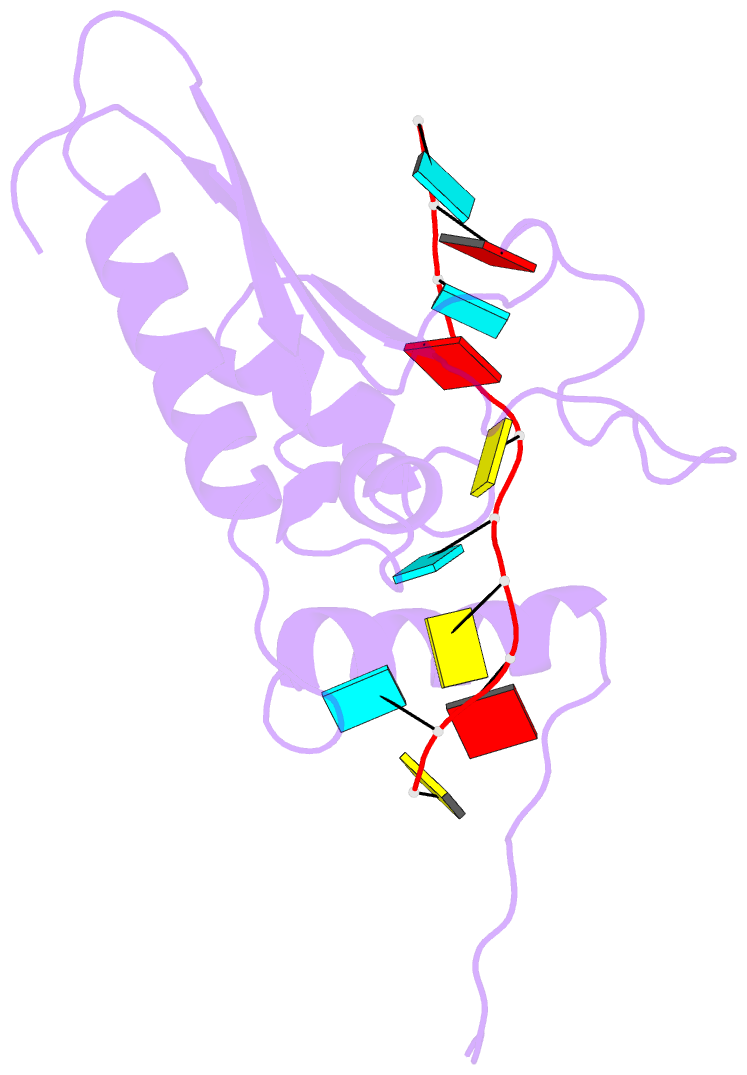
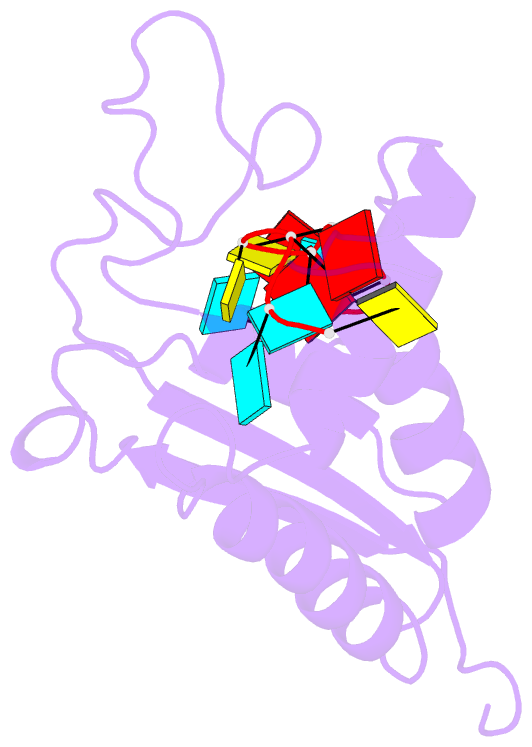
Summary information and primary citation
- PDB-id
- 2mjh; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- NMR
- Summary
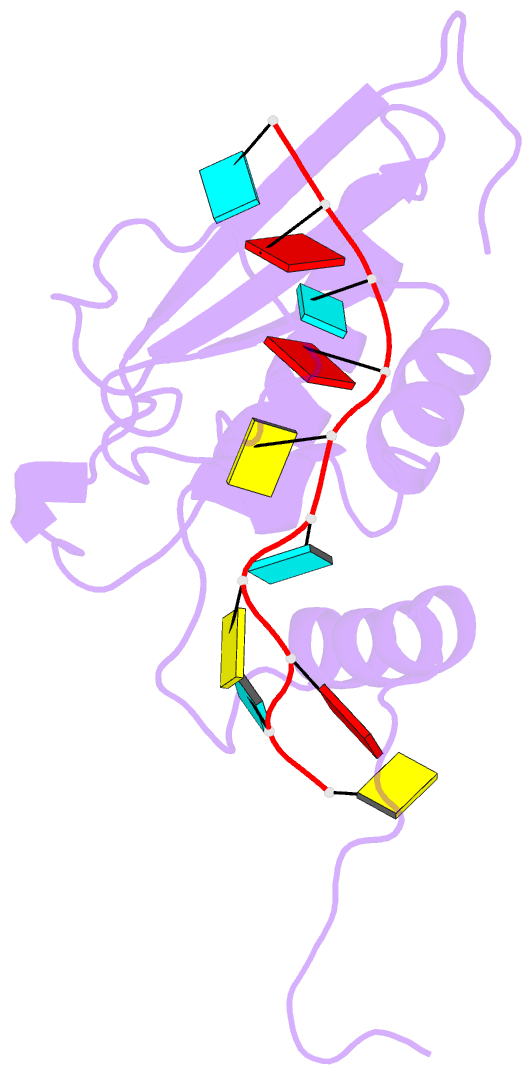
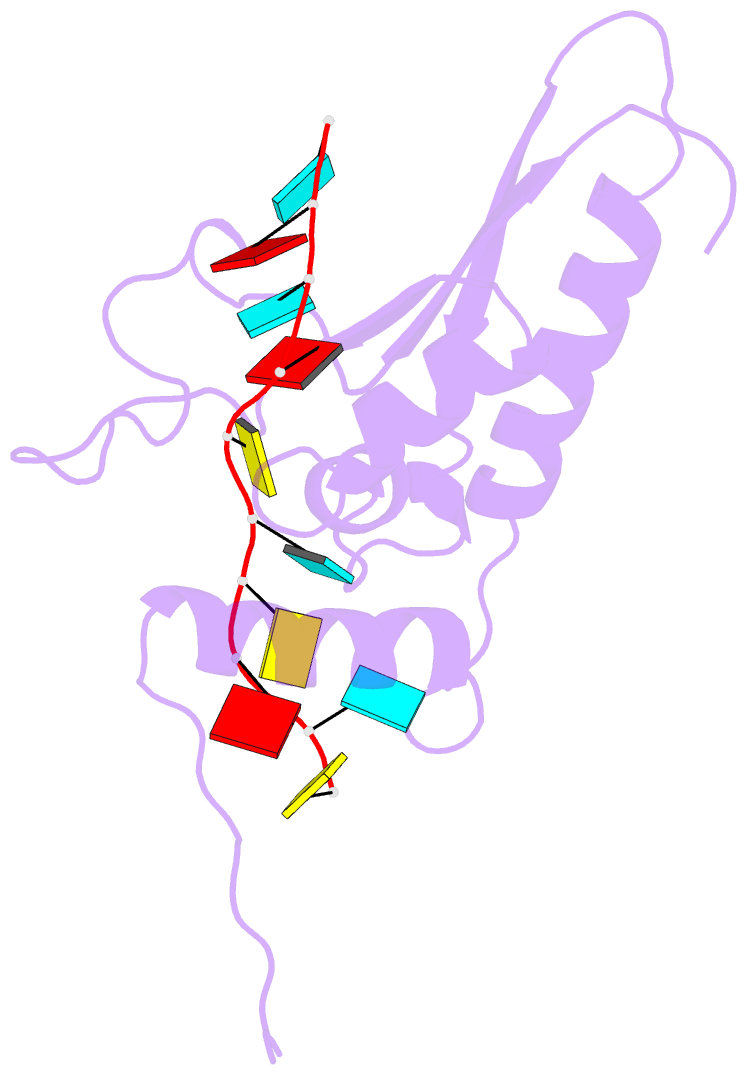
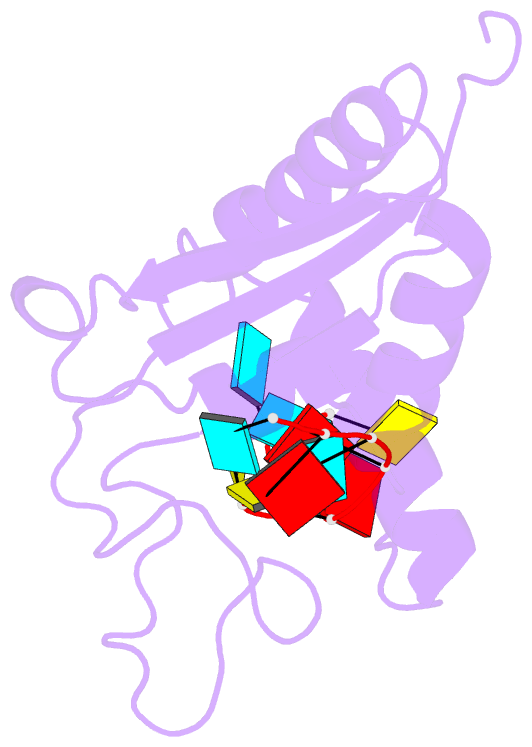
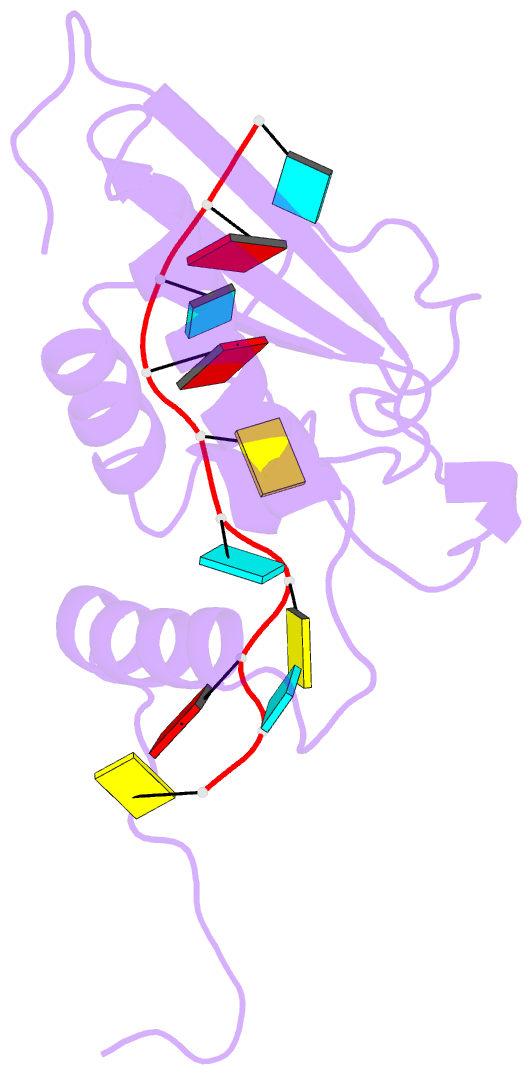
- Solution structure of the gld-1 RNA-binding domain in complex with RNA
- Reference
- Daubner GM, Brummer A, Tocchini C, Gerhardy S, Ciosk R, Zavolan M, Allain FH (2014): "Structural and functional implications of the QUA2 domain on RNA recognition by GLD-1." Nucleic Acids Res., 42, 8092-8105. doi: 10.1093/nar/gku445.
- Abstract
- The STAR family comprises ribonucleic acid (RNA)-binding proteins that play key roles in RNA-regulatory processes. RNA recognition is achieved by a KH domain with an additional α-helix (QUA2) that seems to extend the RNA-binding surface to six nucleotides for SF1 (Homo sapiens) and seven nucleotides for GLD-1 (Caenorhabditis elegans). To understand the structural basis of this probable difference in specificity, we determined the solution structure of GLD-1 KH-QUA2 with the complete consensus sequence identified in the tra-2 gene. Compared to SF1, the GLD-1 KH-QUA2 interface adopts a different conformation resulting indeed in an additional sequence-specific binding pocket for a uracil at the 5'end. The functional relevance of this binding pocket is emphasized by our bioinformatics analysis showing that GLD-1 binding sites with this 5'end uracil are more predictive for the functional response of the messenger RNAs to gld-1 knockout. We further reveal the importance of the KH-QUA2 interface in vitro and that its alteration in vivo affects the level of translational repression dependent on the sequence of the GLD-1 binding motif. In conclusion, we demonstrate that the QUA2 domain distinguishes GLD-1 from other members of the STAR family and contributes more generally to the modulation of RNA-binding affinity and specificity of KH domain containing proteins.