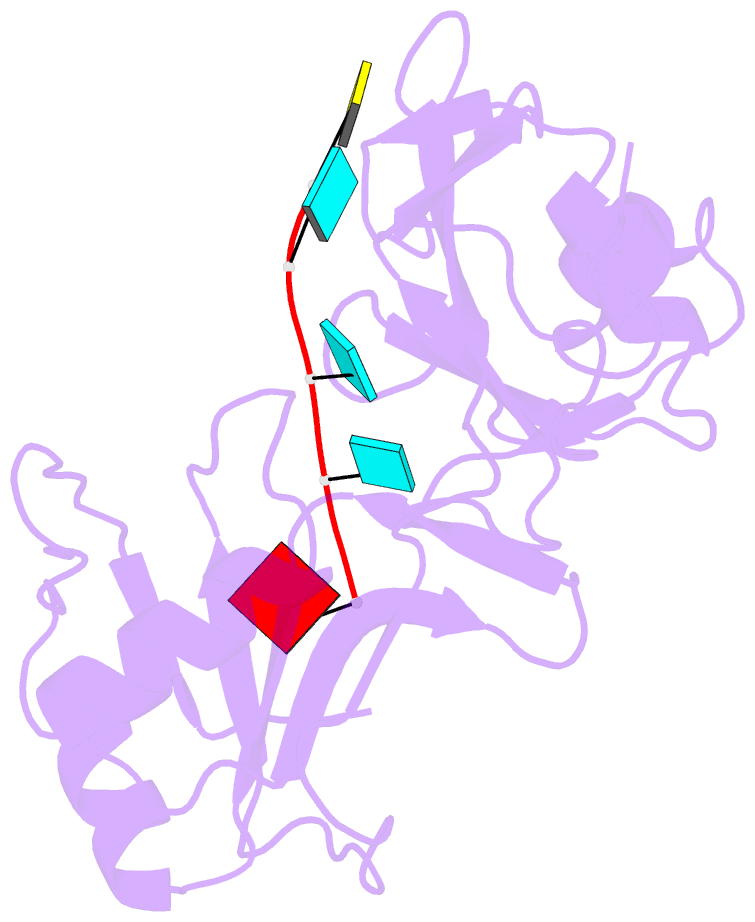
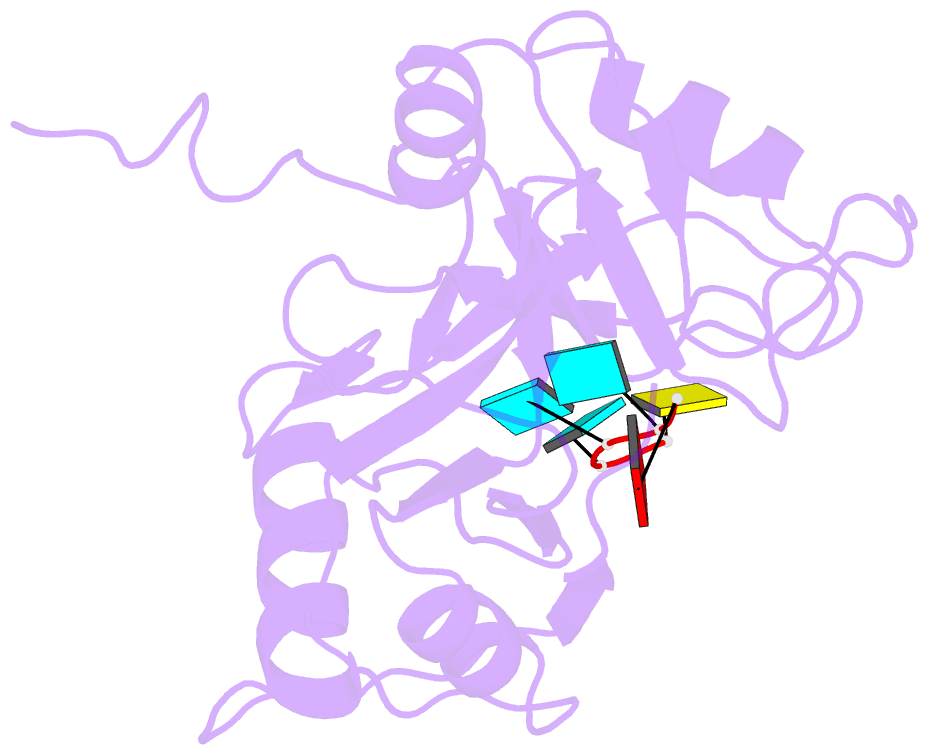
Summary information and primary citation
- PDB-id
- 2mki; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- translation regulator-RNA
- Method
- NMR
- Summary
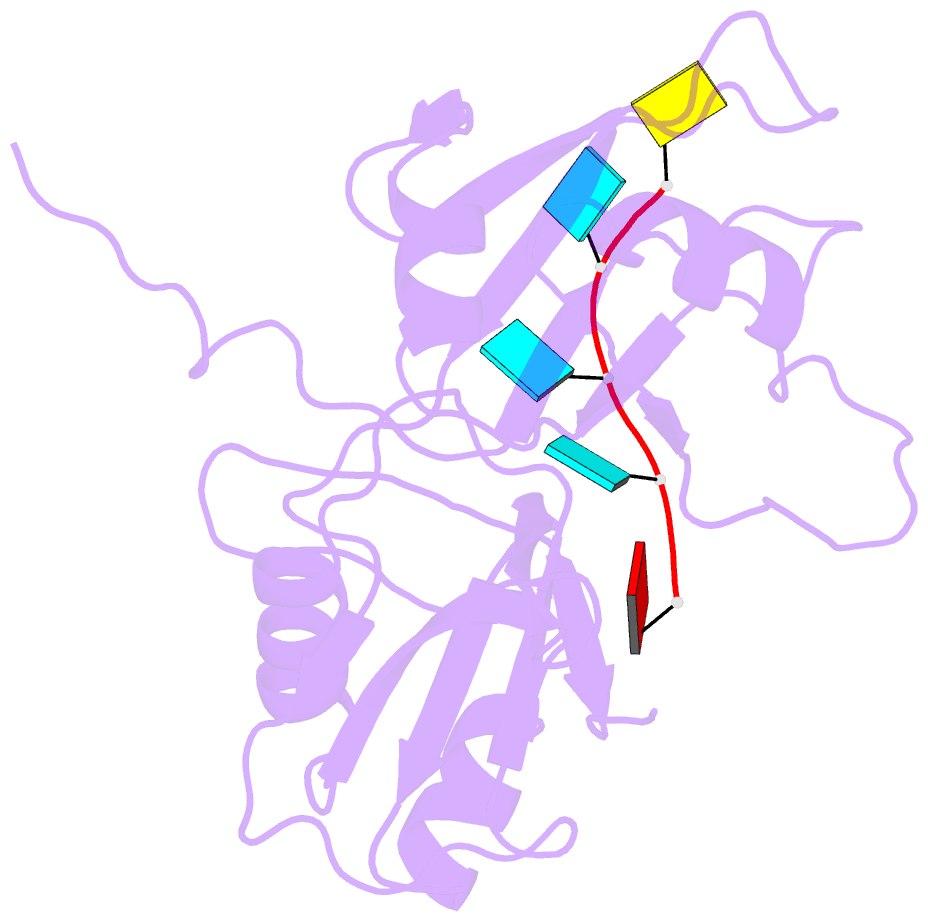
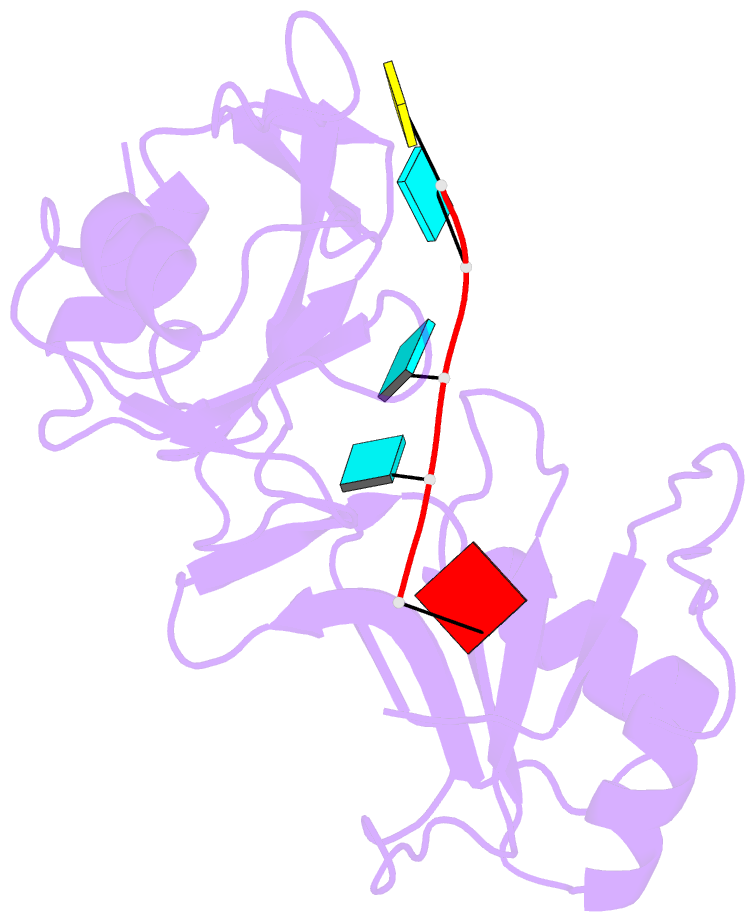
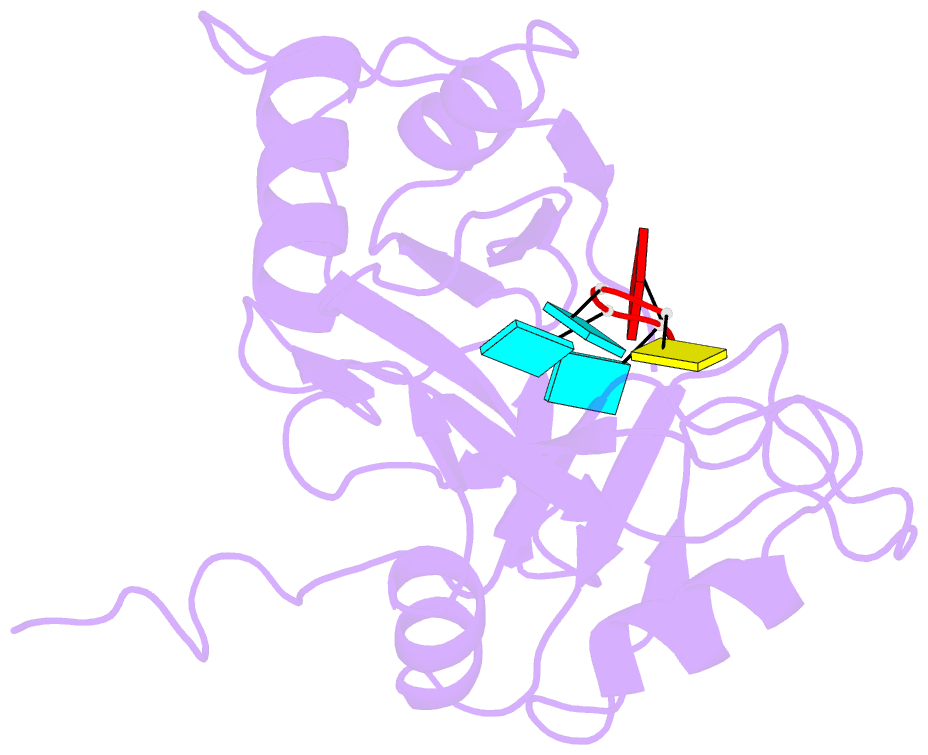
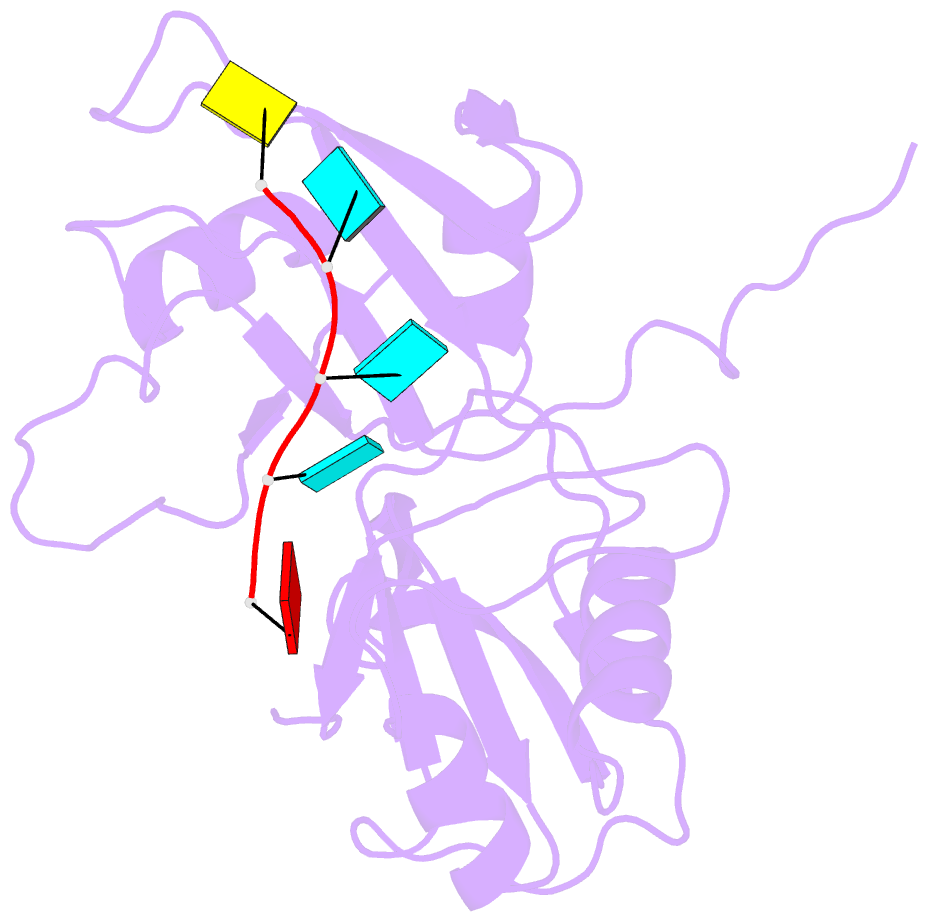
- Solution structure of tandem rrm domains of cytoplasmic polyadenylation element binding protein 4 (cpeb4) in complex with RNA
- Reference
- Afroz T, Skrisovska L, Belloc E, Guillen-Boixet J, Mendez R, Allain FH-T (2014): "A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins." Genes Dev., 28, 1498-1514. doi: 10.1101/gad.241133.114.
- Abstract
- Cytoplasmic changes in polyA tail length is a key mechanism of translational control and is implicated in germline development, synaptic plasticity, cellular proliferation, senescence, and cancer progression. The presence of a U-rich cytoplasmic polyadenylation element (CPE) in the 3' untranslated regions (UTRs) of the responding mRNAs gives them the selectivity to be regulated by the CPE-binding (CPEB) family of proteins, which recognizes RNA via the tandem RNA recognition motifs (RRMs). Here we report the solution structures of the tandem RRMs of two human paralogs (CPEB1 and CPEB4) in their free and RNA-bound states. The structures reveal an unprecedented arrangement of RRMs in the free state that undergo an original closure motion upon RNA binding that ensures high fidelity. Structural and functional characterization of the ZZ domain (zinc-binding domain) of CPEB1 suggests a role in both protein-protein and protein-RNA interactions. Together with functional studies, the structures reveal how RNA binding by CPEB proteins leads to an optimal positioning of the N-terminal and ZZ domains at the 3' UTR, which favors the nucleation of the functional ribonucleoprotein complexes for translation regulation.