Summary information and primary citation
- PDB-id
- 2mxy; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- NMR
- Summary
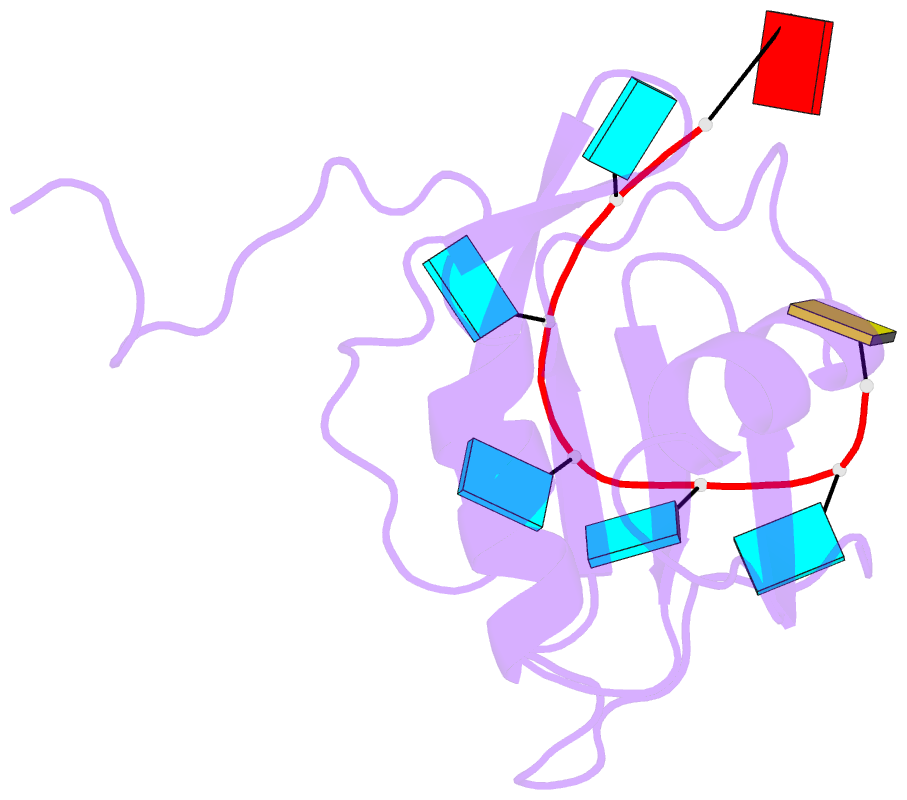
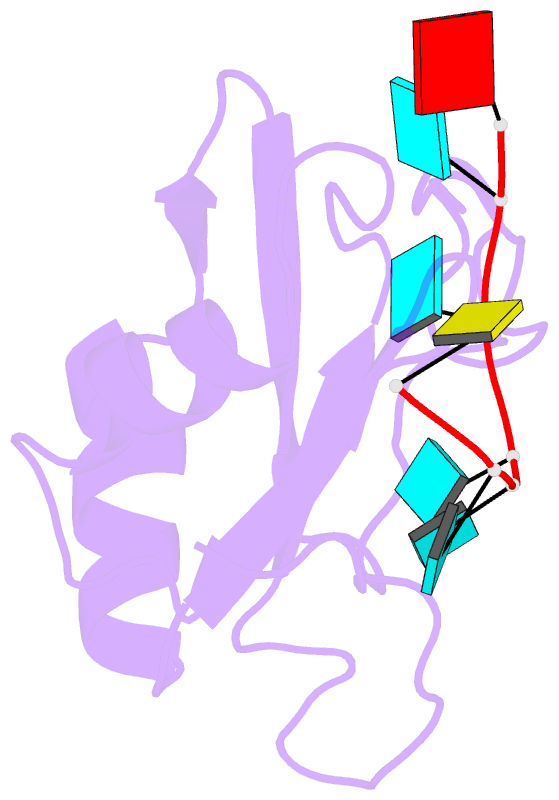
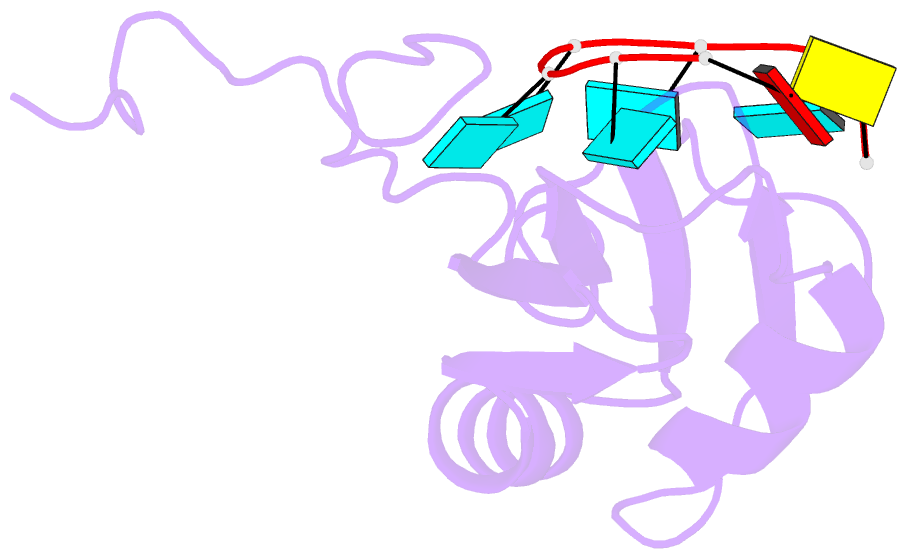
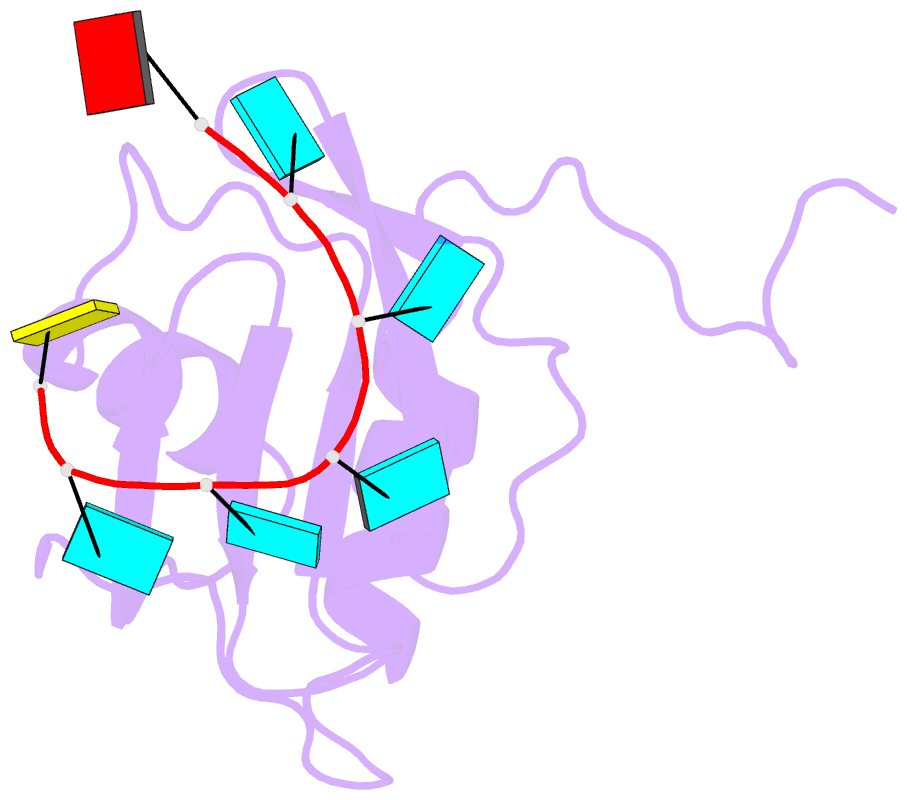
- Solution structure of hnrnp c rrm in complex with 5'-auuuuuc-3' RNA
- Reference
- Cienikova Z, Damberger FF, Hall J, Allain FH, Maris C (2014): "Structural and mechanistic insights into poly(uridine) tract recognition by the hnRNP C RNA recognition motif." J.Am.Chem.Soc., 136, 14536-14544. doi: 10.1021/ja507690d.
- Abstract
- HnRNP C is a ubiquitous RNA regulatory factor and the principal constituent of the nuclear hnRNP core particle. The protein contains one amino-terminal RNA recognition motif (RRM) known to bind uridine (U)-rich sequences. This work provides a molecular and mechanistic understanding of this interaction. We solved the solution structures of the RRM in complex with poly(U) oligomers of five and seven nucleotides. The five binding pockets of RRM recognize uridines with an unusual 5'-to-3' gradient of base selectivity. The target recognition is therefore strongly sensitive to base clustering, explaining the preference for contiguous uridine tracts. Using a novel approach integrating the structurally derived recognition consensus of the RRM with a thermodynamic description of its multi-register binding, we modeled the saturation of cellular uridine tracts by this protein. The binding pattern is remarkably consistent with the experimentally observed transcriptome-wide cross-link distribution of the full-length hnRNP C on short uridine tracts. This result re-establishes the RRM as the primary RNA-binding domain of the hnRNP C tetramer and provides a proof of concept for interpreting high-throughput interaction data using structural approaches.