Summary information and primary citation
- PDB-id
- 2nqj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (2.45 Å)
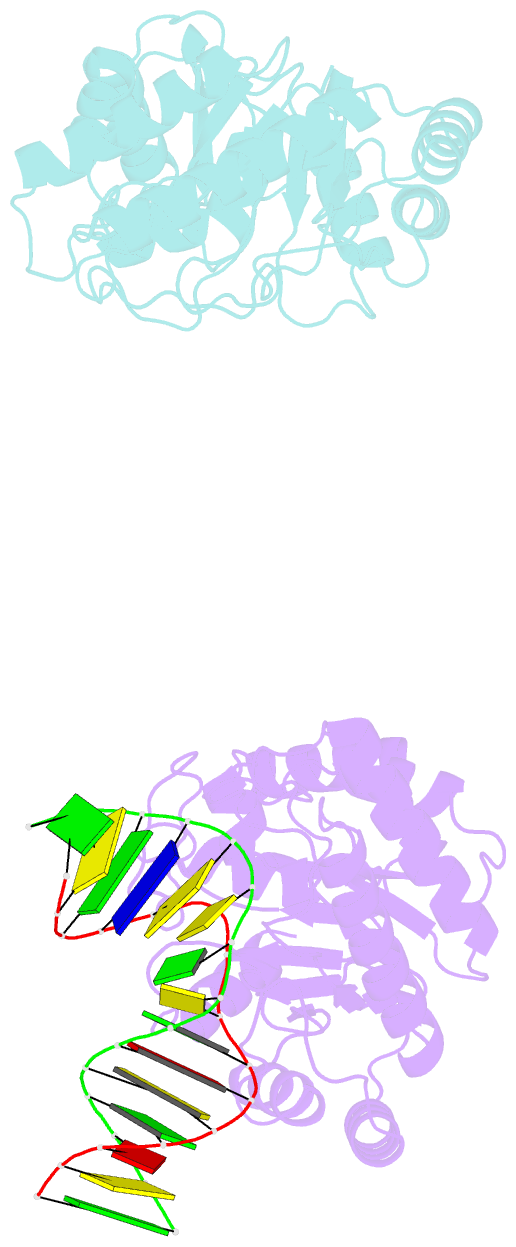
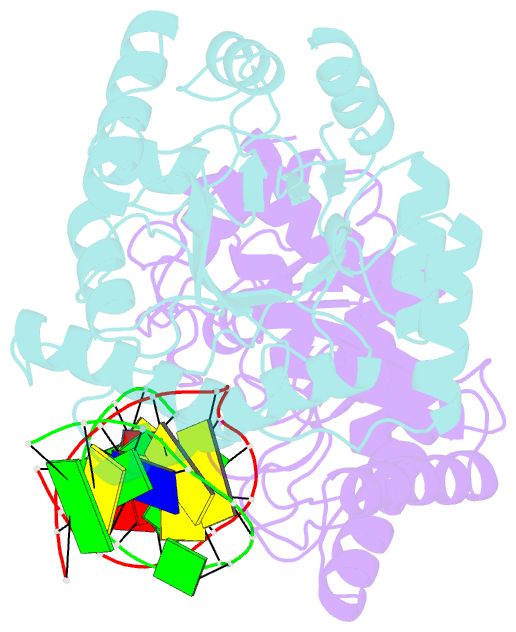
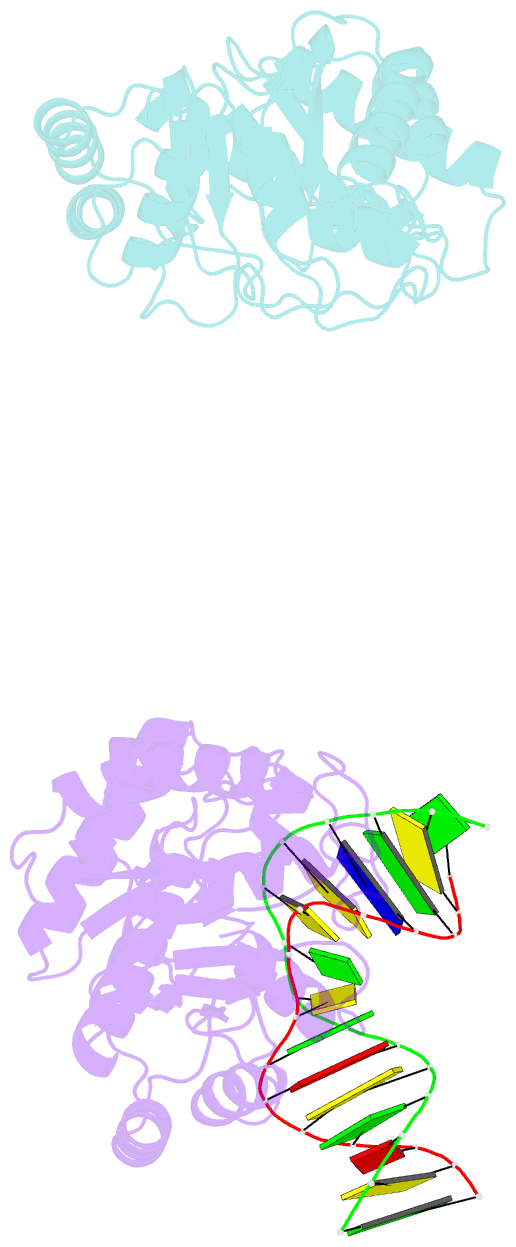
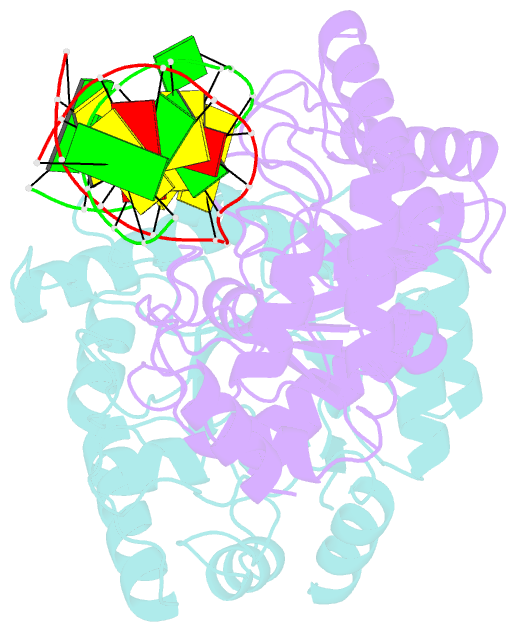
- Summary
- Crystal structure of escherichia coli endonuclease iv (endo iv) e261q mutant bound to damaged DNA
- Reference
- Garcin ED, Hosfield DJ, Desai SA, Haas BJ, Bjoras M, Cunningham RP, Tainer JA (2008): "DNA apurinic-apyrimidinic site binding and excision by endonuclease IV." Nat.Struct.Mol.Biol., 15, 515-522. doi: 10.1038/nsmb.1414.
- Abstract
- Escherichia coli endonuclease IV is an archetype for an abasic or apurinic-apyrimidinic endonuclease superfamily crucial for DNA base excision repair. Here biochemical, mutational and crystallographic characterizations reveal a three-metal ion mechanism for damage binding and incision. The 1.10-A resolution DNA-free and the 2.45-A resolution DNA-substrate complex structures capture substrate stabilization by Arg37 and reveal a distorted Zn3-ligand arrangement that reverts, after catalysis, to an ideal geometry suitable to hold rather than release cleaved DNA product. The 1.45-A resolution DNA-product complex structure shows how Tyr72 caps the active site, tunes its dielectric environment and promotes catalysis by Glu261-activated hydroxide, bound to two Zn2+ ions throughout catalysis. These structural, mutagenesis and biochemical results suggest general requirements for abasic site removal in contrast to features specific to the distinct endonuclease IV alpha-beta triose phosphate isomerase (TIM) barrel and APE1 four-layer alpha-beta folds of the apurinic-apyrimidinic endonuclease families.