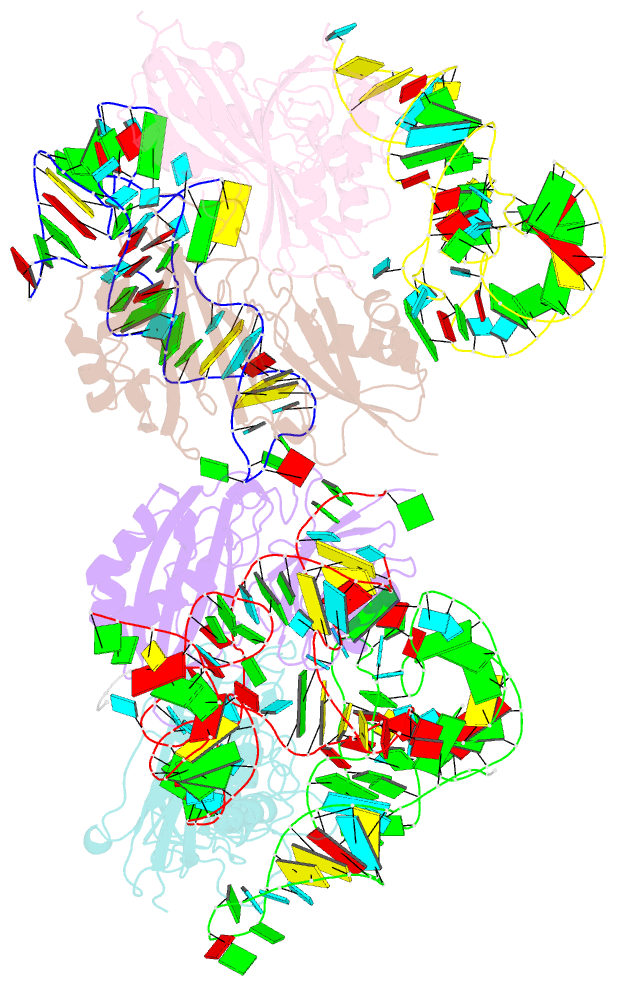
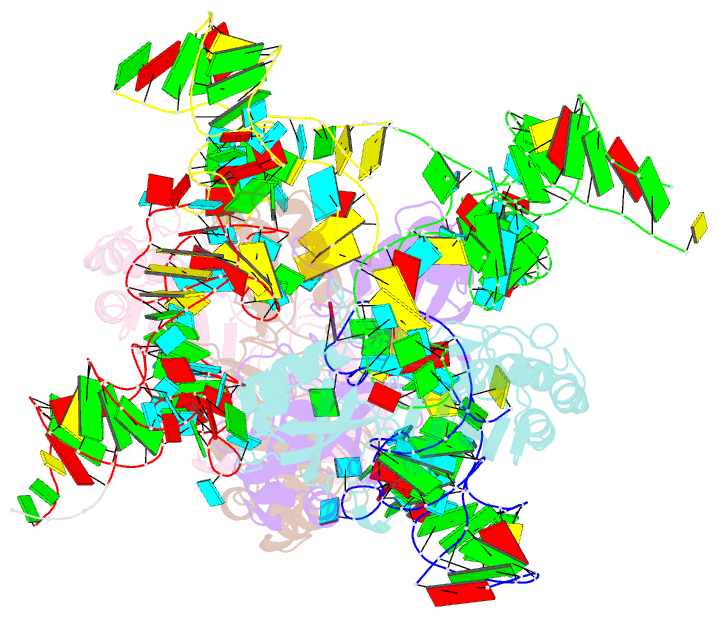
Summary information and primary citation
- PDB-id
- 2nr0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase-RNA
- Method
- X-ray (3.9 Å)
- Summary
- Crystal structure of pseudoudirinde synthase trua in complex with leucyl trna
- Reference
- Hur S, Stroud RM (2007): "How U38, 39, and 40 of Many tRNAs Become the Targets for Pseudouridylation by TruA." Mol.Cell, 26, 189-203. doi: 10.1016/j.molcel.2007.02.027.
- Abstract
- Translational accuracy and efficiency depend upon modification of uridines in the tRNA anticodon stem loop (ASL) by a highly conserved pseudouridine synthase TruA. TruA specifically modifies uridines at positions 38, 39, and/or 40 of tRNAs with highly divergent sequences and structures through a poorly characterized mechanism that differs from previously studied RNA-modifying enzymes. The molecular basis for the site and substrate "promiscuity" was studied by determining the crystal structures of E. coli TruA in complex with two different leucyl tRNAs in conjunction with functional assays and computer simulation. The structures capture three stages of the TruA*tRNA reaction, revealing the mechanism by which TruA selects the target site. We propose that TruA utilizes the intrinsic flexibility of the ASL for site promiscuity and also to select against intrinsically stable tRNAs to avoid their overstabilization through pseudouridylation, thereby maintaining the balance between the flexibility and stability required for its biological function.