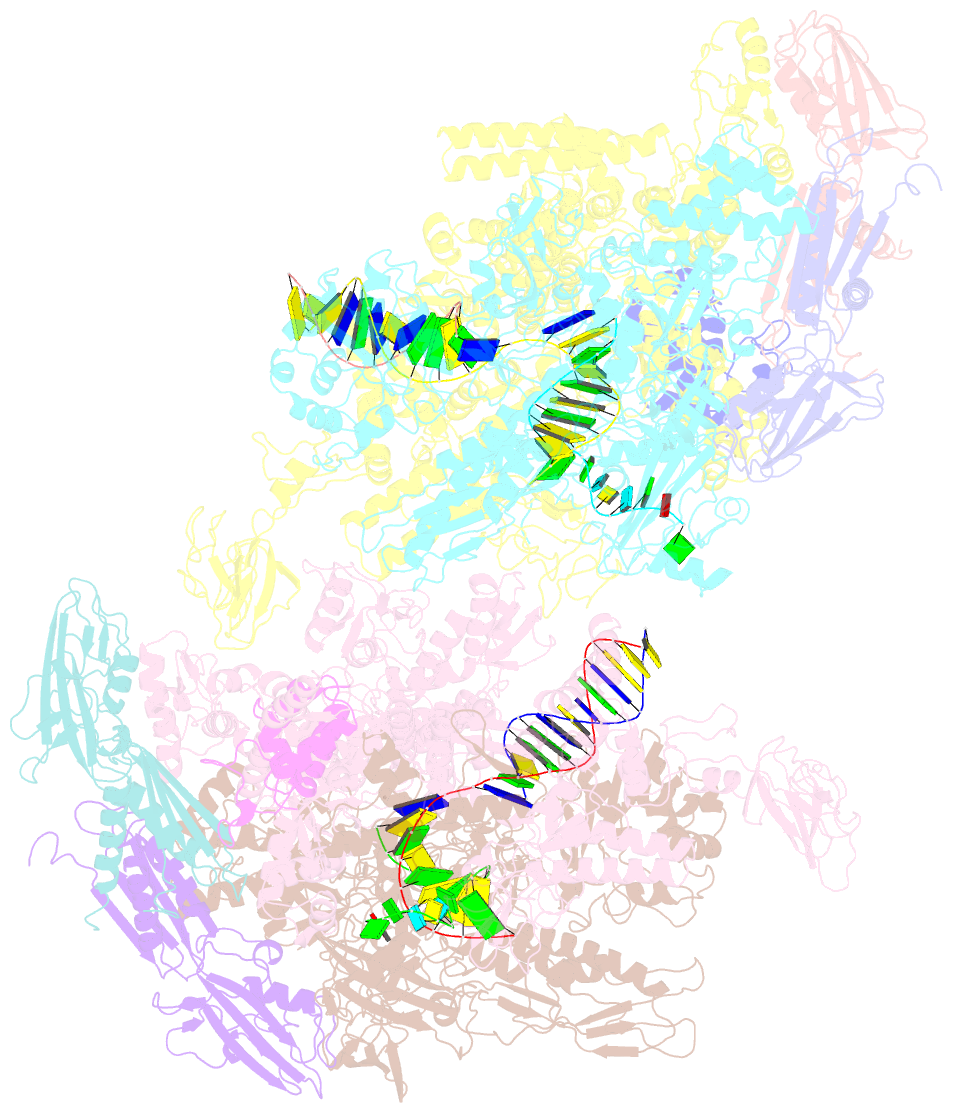
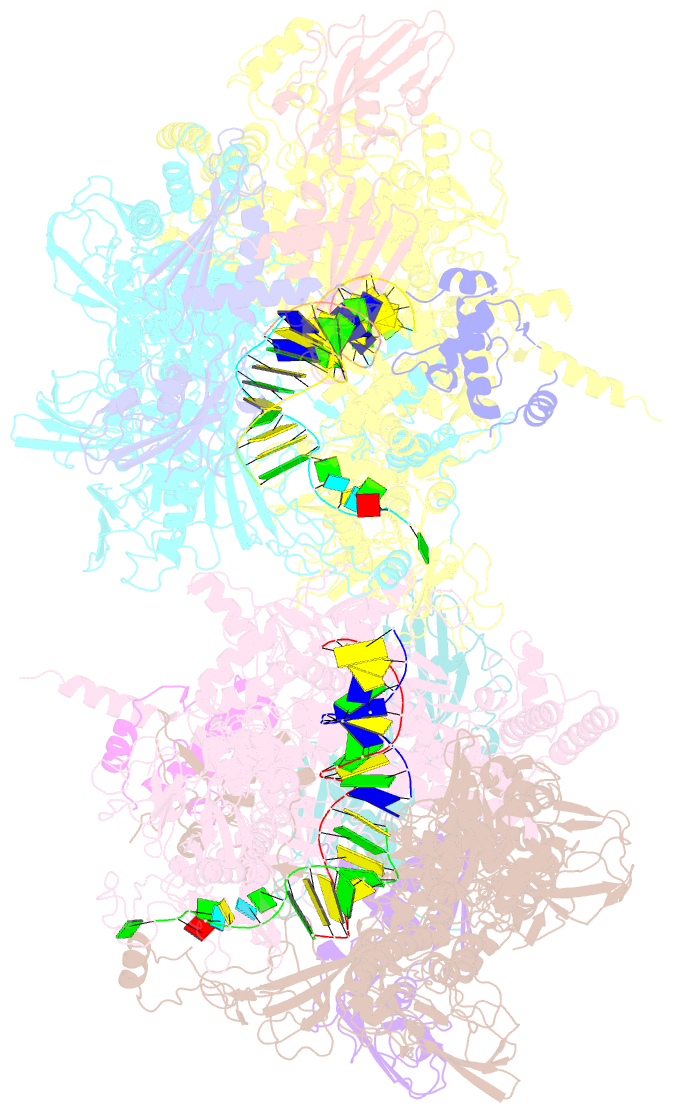
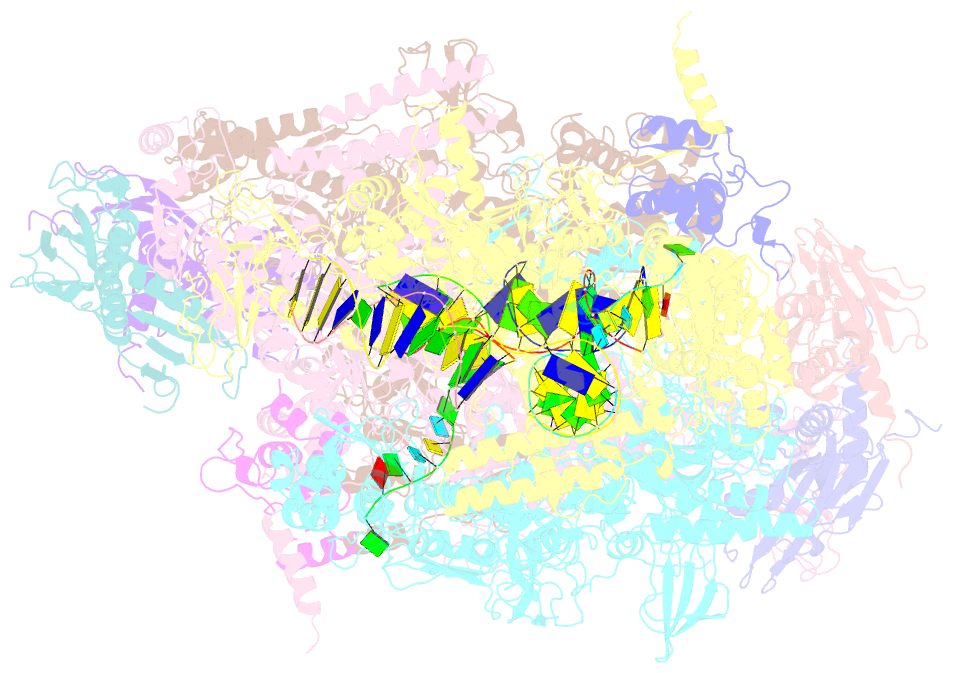
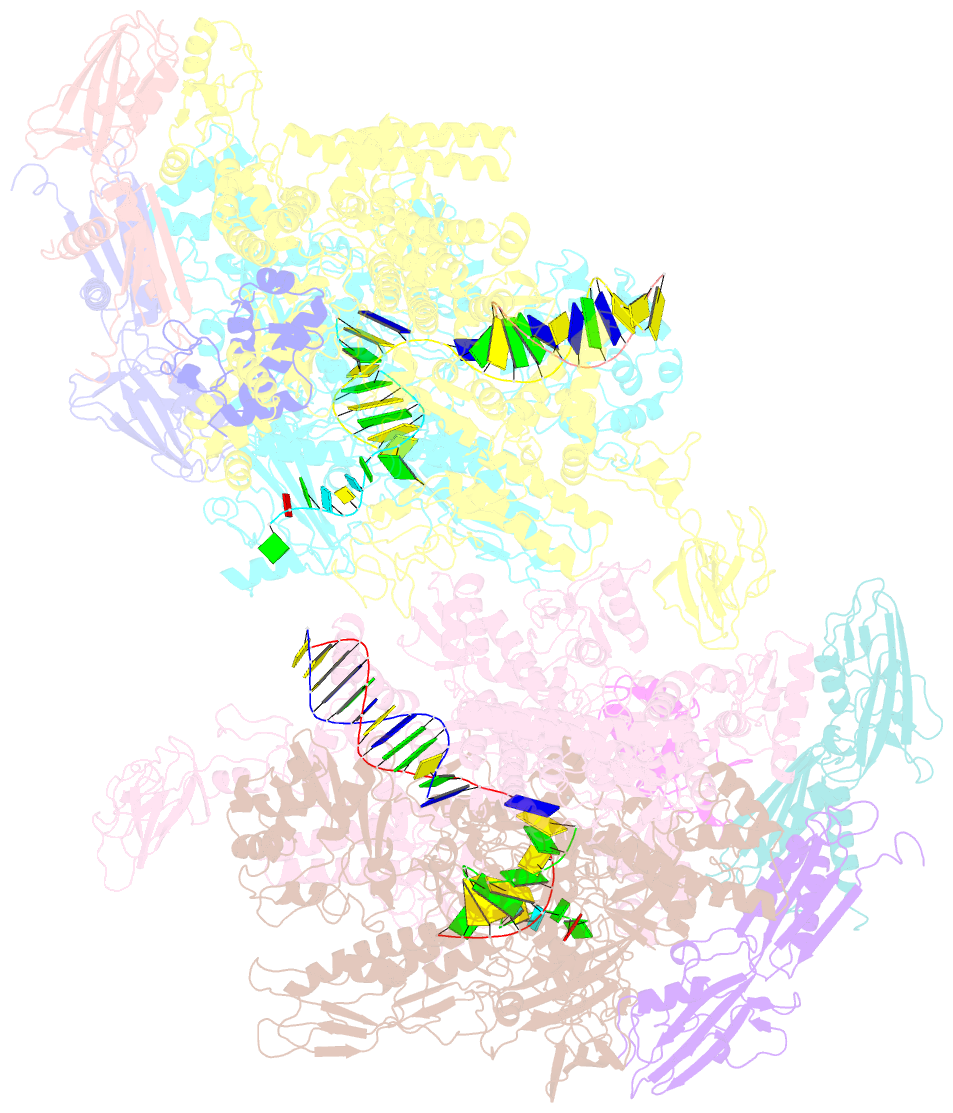
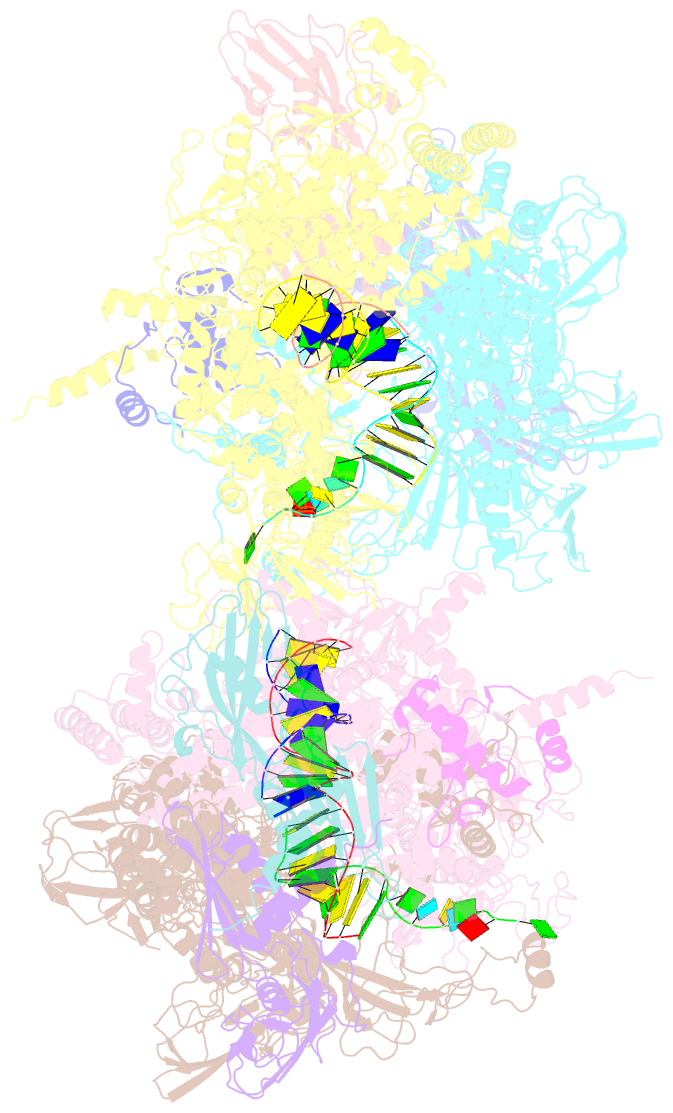
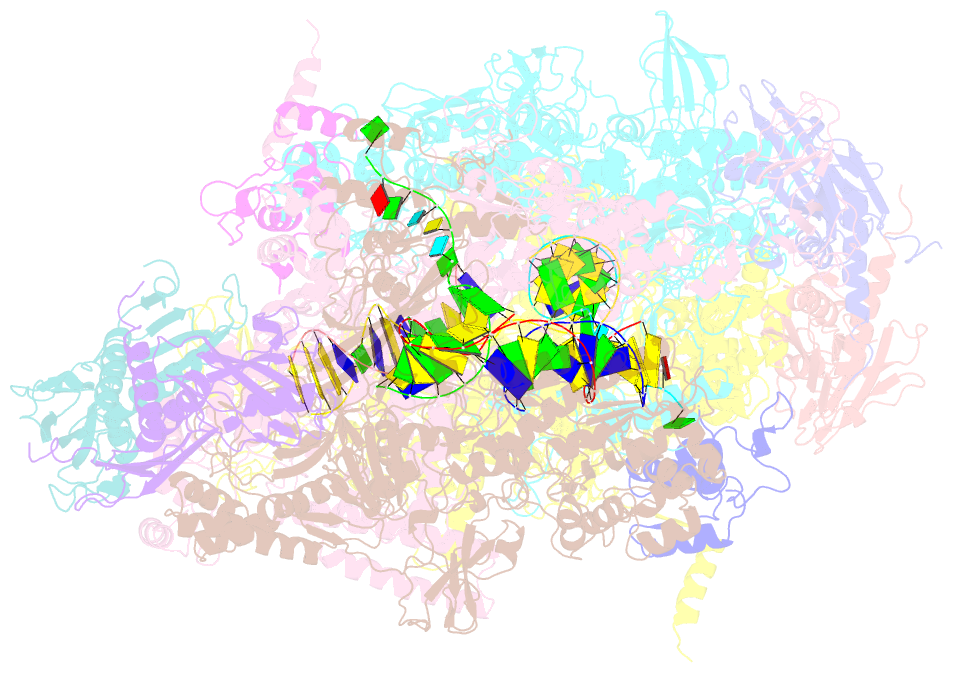
Summary information and primary citation
- PDB-id
- 2o5j; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA-RNA hybrid
- Method
- X-ray (3.0 Å)
- Summary
- Crystal structure of the t. thermophilus rnap polymerase elongation complex with the ntp substrate analog
- Reference
- Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R (2007): "Structural basis for substrate loading in bacterial RNA polymerase." Nature, 448, 163-168. doi: 10.1038/nature05931.
- Abstract
- The mechanism of substrate loading in multisubunit RNA polymerase is crucial for understanding the general principles of transcription yet remains hotly debated. Here we report the 3.0-A resolution structures of the Thermus thermophilus elongation complex (EC) with a non-hydrolysable substrate analogue, adenosine-5'-[(alpha,beta)-methyleno]-triphosphate (AMPcPP), and with AMPcPP plus the inhibitor streptolydigin. In the EC/AMPcPP structure, the substrate binds to the active ('insertion') site closed through refolding of the trigger loop (TL) into two alpha-helices. In contrast, the EC/AMPcPP/streptolydigin structure reveals an inactive ('preinsertion') substrate configuration stabilized by streptolydigin-induced displacement of the TL. Our structural and biochemical data suggest that refolding of the TL is vital for catalysis and have three main implications. First, despite differences in the details, the two-step preinsertion/insertion mechanism of substrate loading may be universal for all RNA polymerases. Second, freezing of the preinsertion state is an attractive target for the design of novel antibiotics. Last, the TL emerges as a prominent target whose refolding can be modulated by regulatory factors.