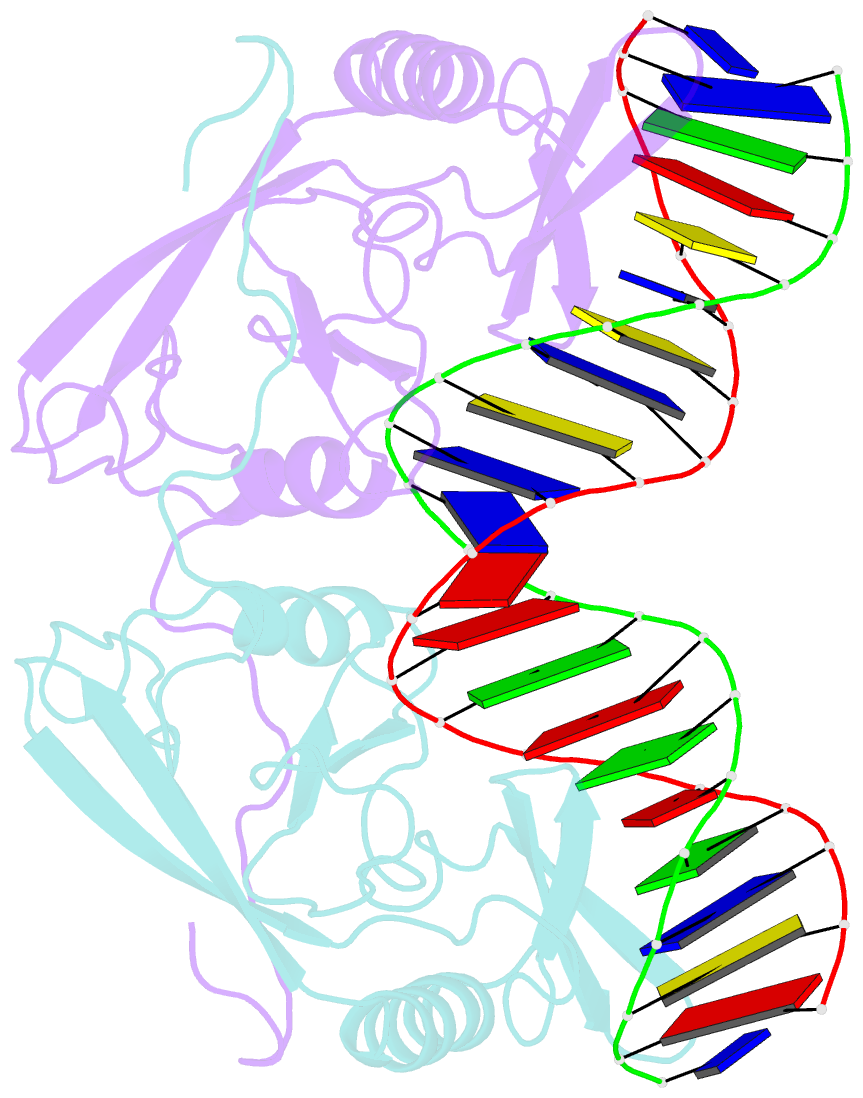
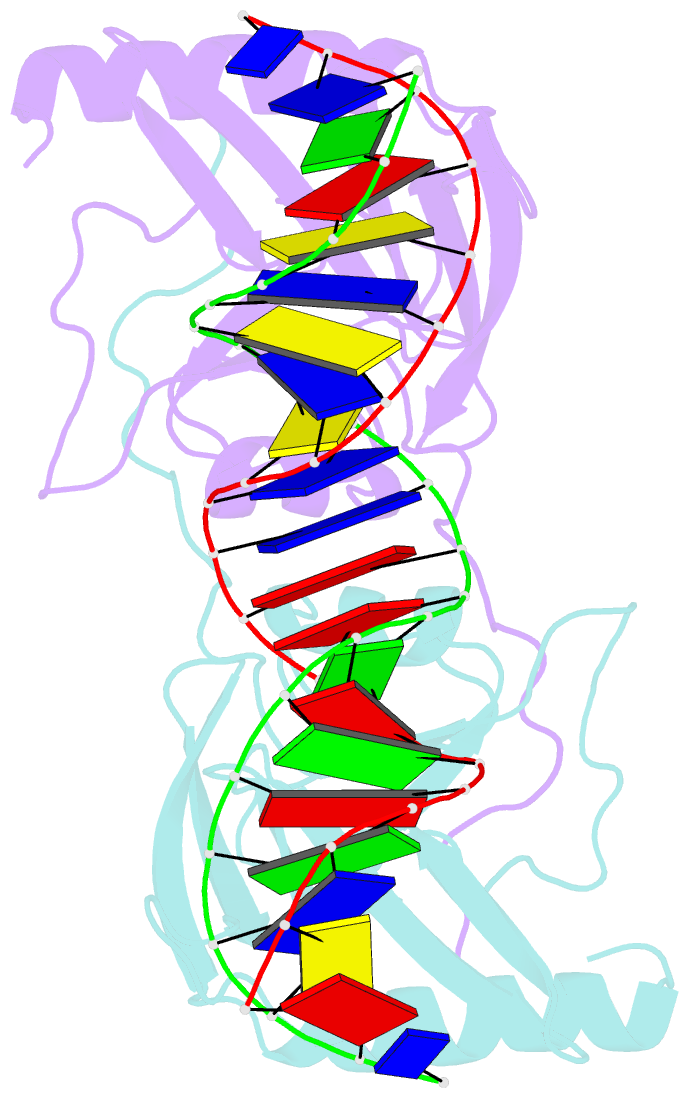
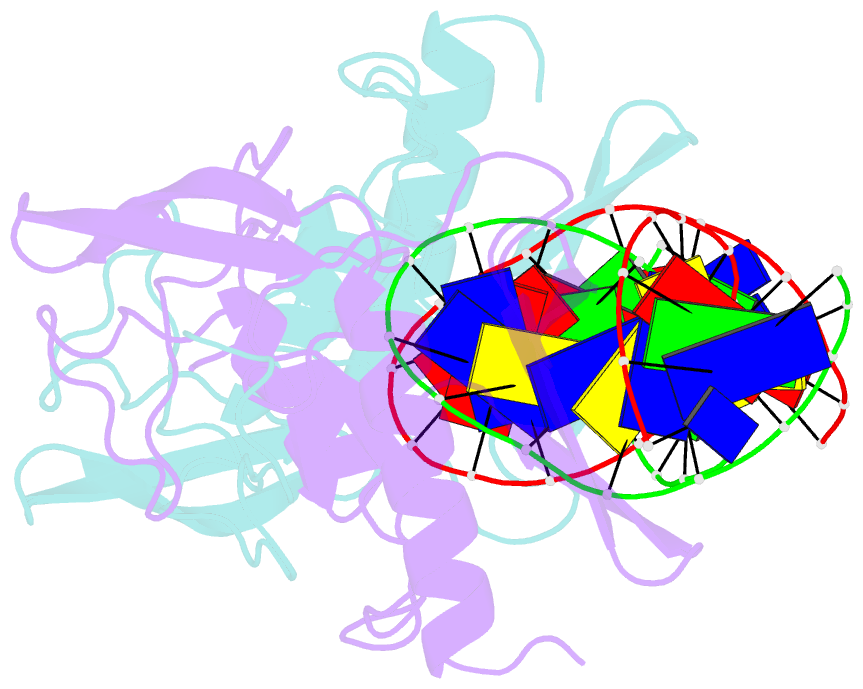
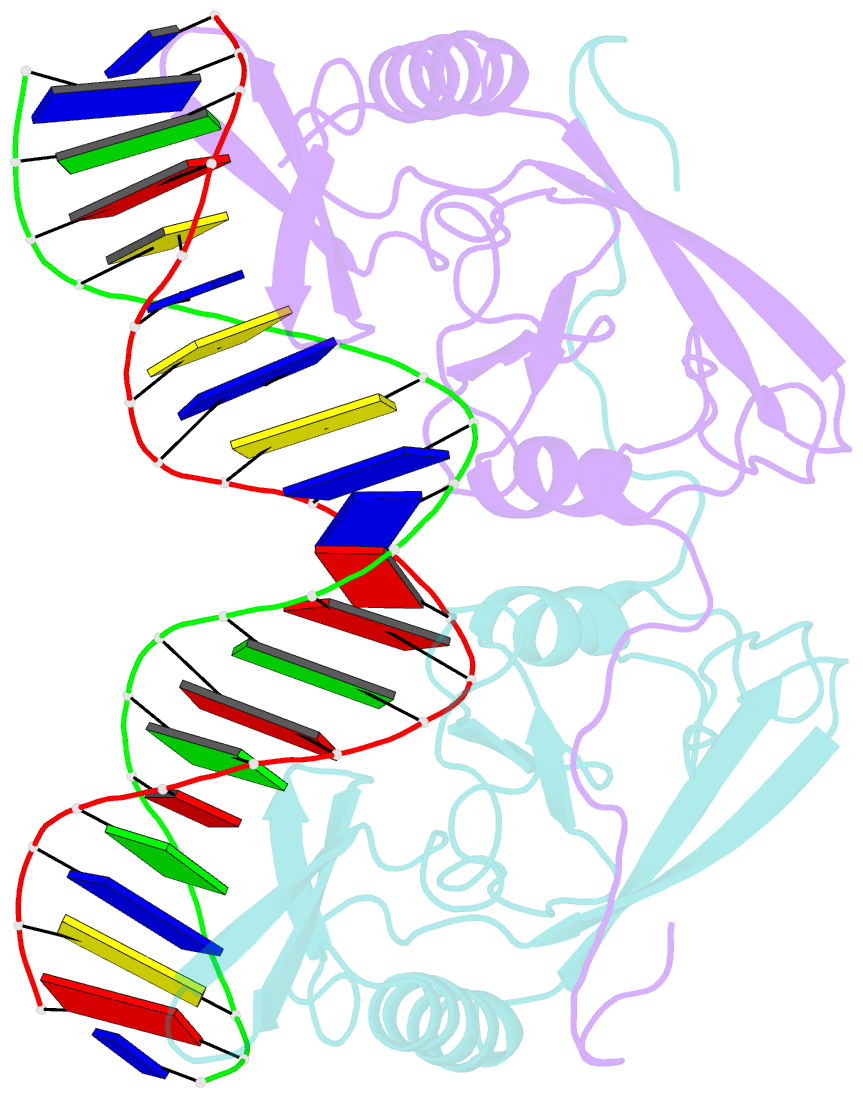
Summary information and primary citation
- PDB-id
- 2o6m; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.3 Å)
- Summary
- H98q mutant of the homing endonuclease i-ppoi complexed with DNA
- Reference
- Eastberg JH, Eklund J, Monnat R, Stoddard BL (2007): "Mutability of an HNH nuclease imidazole general base and exchange of a deprotonation mechanism." Biochemistry, 46, 7215-7225. doi: 10.1021/bi700418d.
- Abstract
- Several unique protein folds that catalyze the hydrolysis of phosphodiester bonds have arisen independently in nature, including the PD(D/E)XK superfamily (typified by type II restriction endonucleases and many recombination and repair enzymes) and the HNH superfamily (found in an equally wide array of enzymes, including bacterial colicins and homing endonucleases). Whereas the identity and position of catalytic residues within the PD(D/E)XK superfamily are highly variable, the active sites of HNH nucleases are much more strongly conserved. In this study, the ability of an HNH nuclease to tolerate a mutation of its most conserved catalytic residue (its histidine general base), and the mechanism of the most active enzyme variant, were characterized. Conversion of this residue into several altered chemistries, glutamine, lysine, or glutamate, resulted in measurable activity. The histidine to glutamine mutant displays the highest residual activity and a pH profile similar to that of the wild-type enzyme. This activity is dependent on the presence of a neighboring imidazole ring, which has taken over as a less efficient general base for the reaction. This result implies that mutational pathways to alternative HNH-derived catalytic sites do exist but are not as extensively or successfully diverged or reoptimized in nature as variants of the PD(D/E)XK nuclease superfamily. This is possibly due to multiple steric constraints placed on the compact HNH motif, which is simultaneously involved in protein folding, DNA binding, and catalysis, as well as the use of a planar, aromatic imidazole group as a general base.