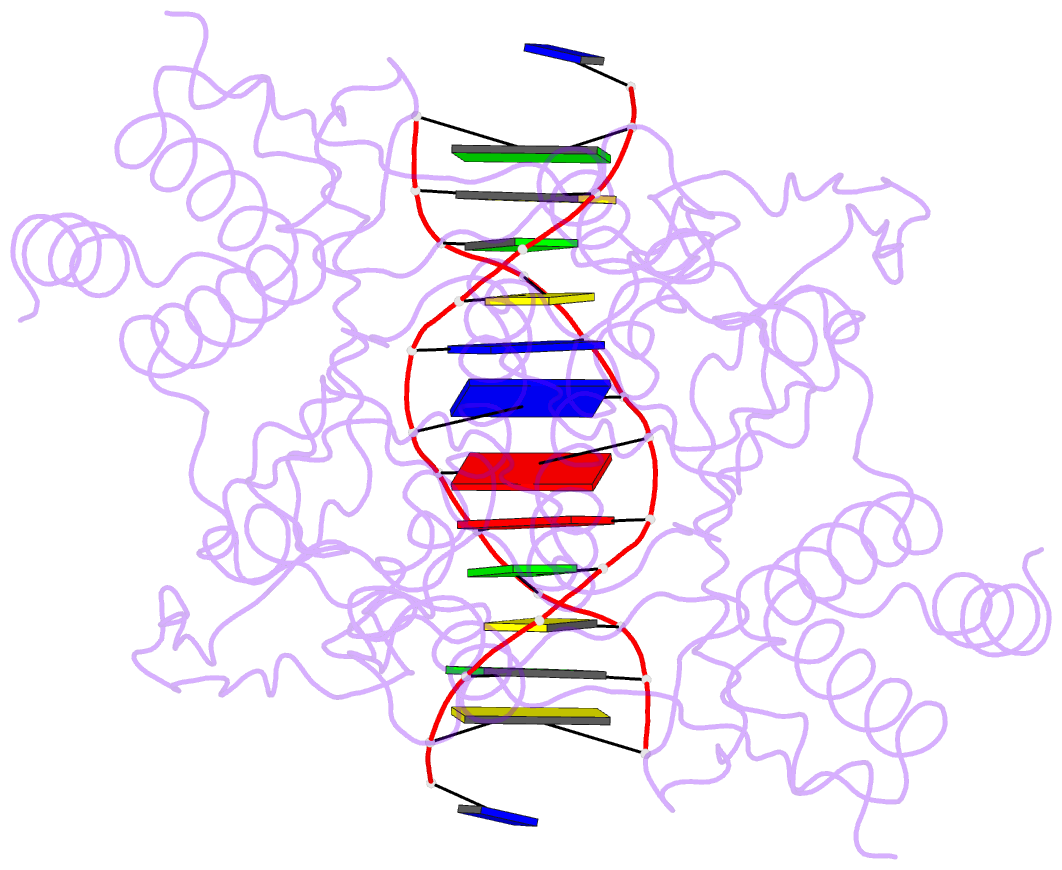
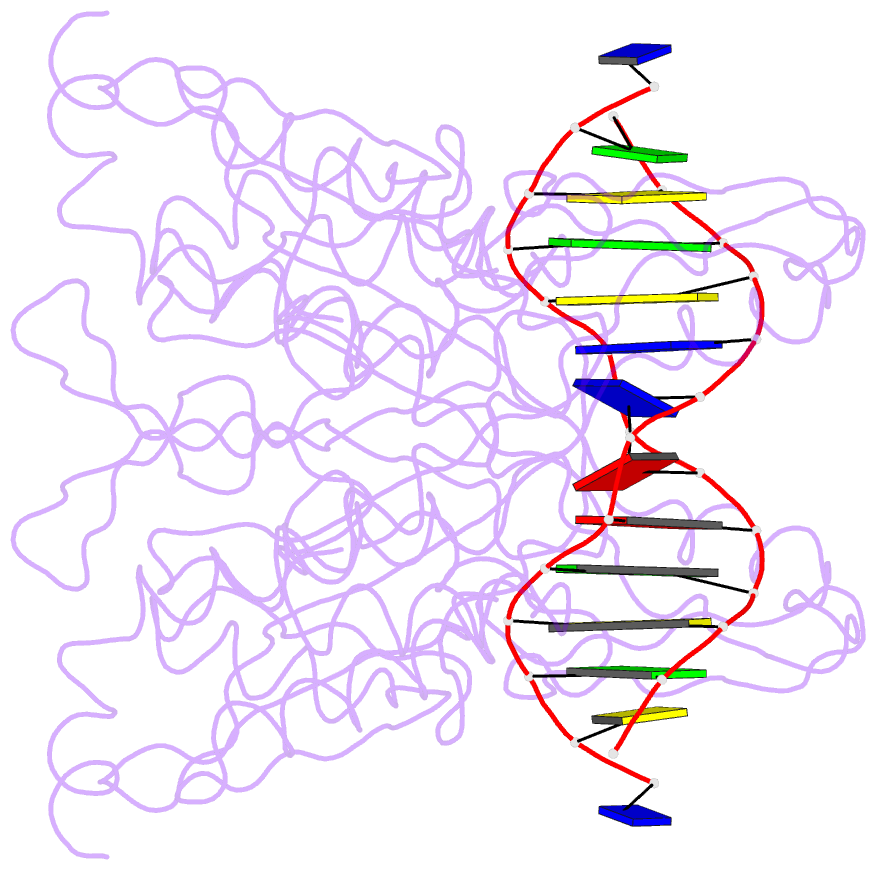
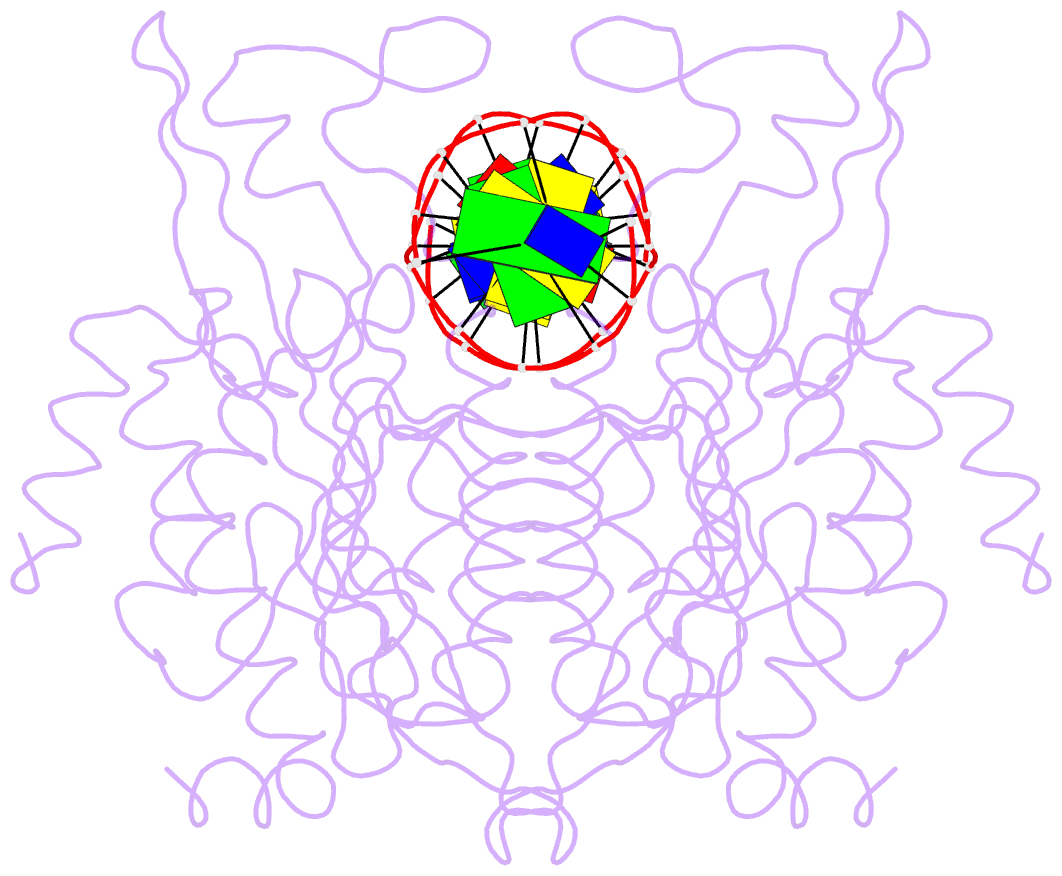
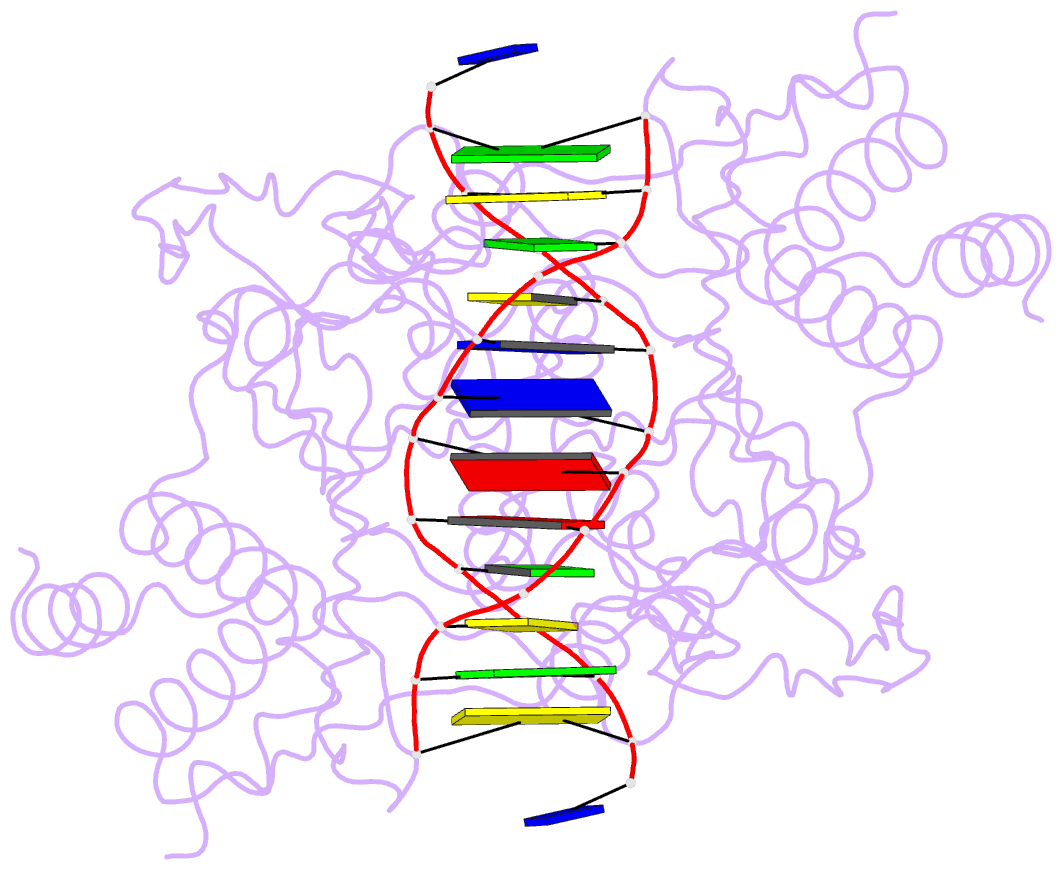
Summary information and primary citation
- PDB-id
- 2oxv; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.95 Å)
- Summary
- Structure of the a138t promiscuous mutant of the ecori restriction endonuclease bound to its cognate recognition site.
- Reference
- Sapienza PJ, Rosenberg JM, Jen-Jacobson L (2007): "Structural and thermodynamic basis for enhanced DNA binding by a promiscuous mutant EcoRI endonuclease." Structure, 15, 1368-1382. doi: 10.1016/j.str.2007.09.014.
- Abstract
- Promiscuous mutant EcoRI endonucleases bind to the canonical site GAATTC more tightly than does the wild-type endonuclease, yet cleave variant (EcoRI(*)) sites more rapidly than does wild-type. The crystal structure of the A138T promiscuous mutant homodimer in complex with a GAATTC site is nearly identical to that of the wild-type complex, except that the Thr138 side chains make packing interactions with bases in the 5'-flanking regions outside the recognition hexanucleotide while excluding two bound water molecules seen in the wild-type complex. Molecular dynamics simulations confirm exclusion of these waters. The structure and simulations suggest possible reasons why binding of the A138T protein to the GAATTC site has DeltaS degrees more favorable and DeltaH degrees less favorable than for wild-type endonuclease binding. The interactions of Thr138 with flanking bases may permit A138T, unlike wild-type enzyme, to form complexes with EcoRI(*) sites that structurally resemble the specific wild-type complex with GAATTC.