Summary information and primary citation
- PDB-id
- 2p66; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase, lyase-DNA
- Method
- X-ray (2.5 Å)
- Summary
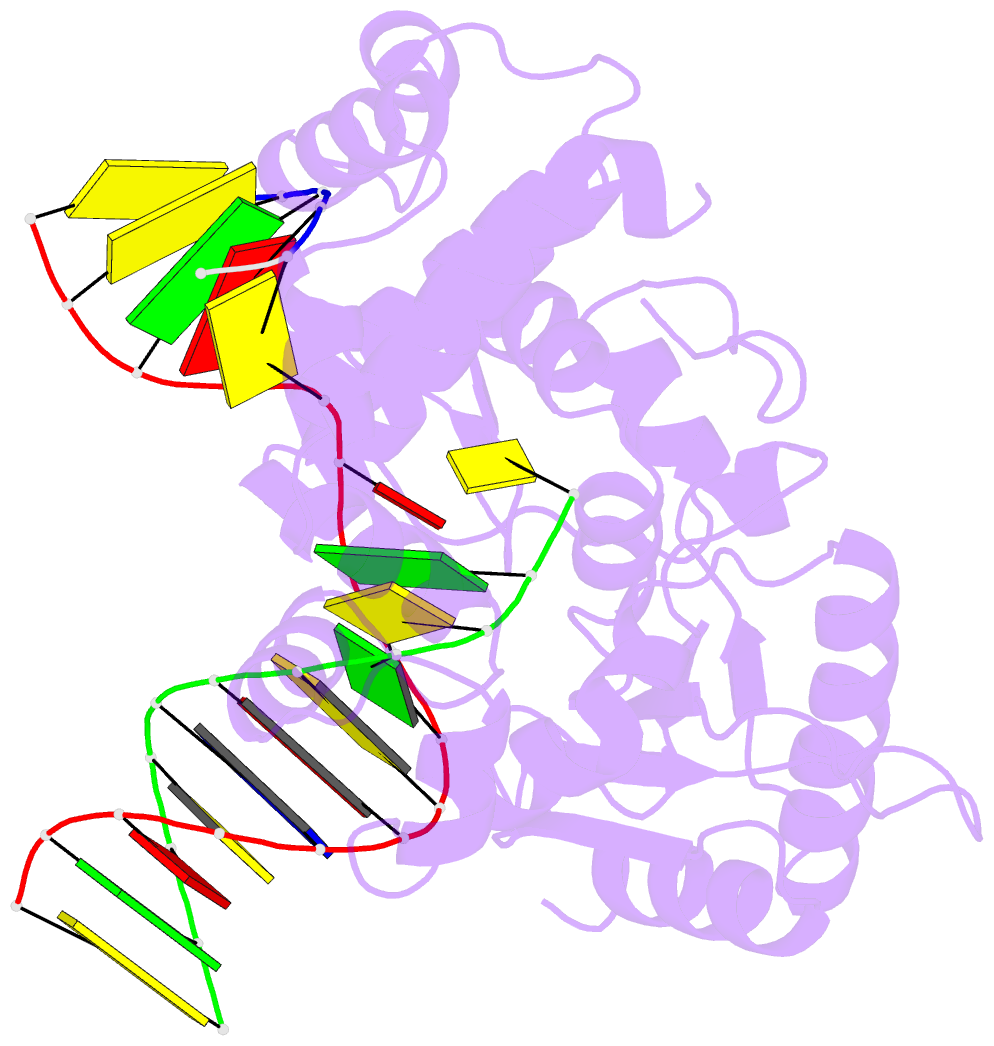
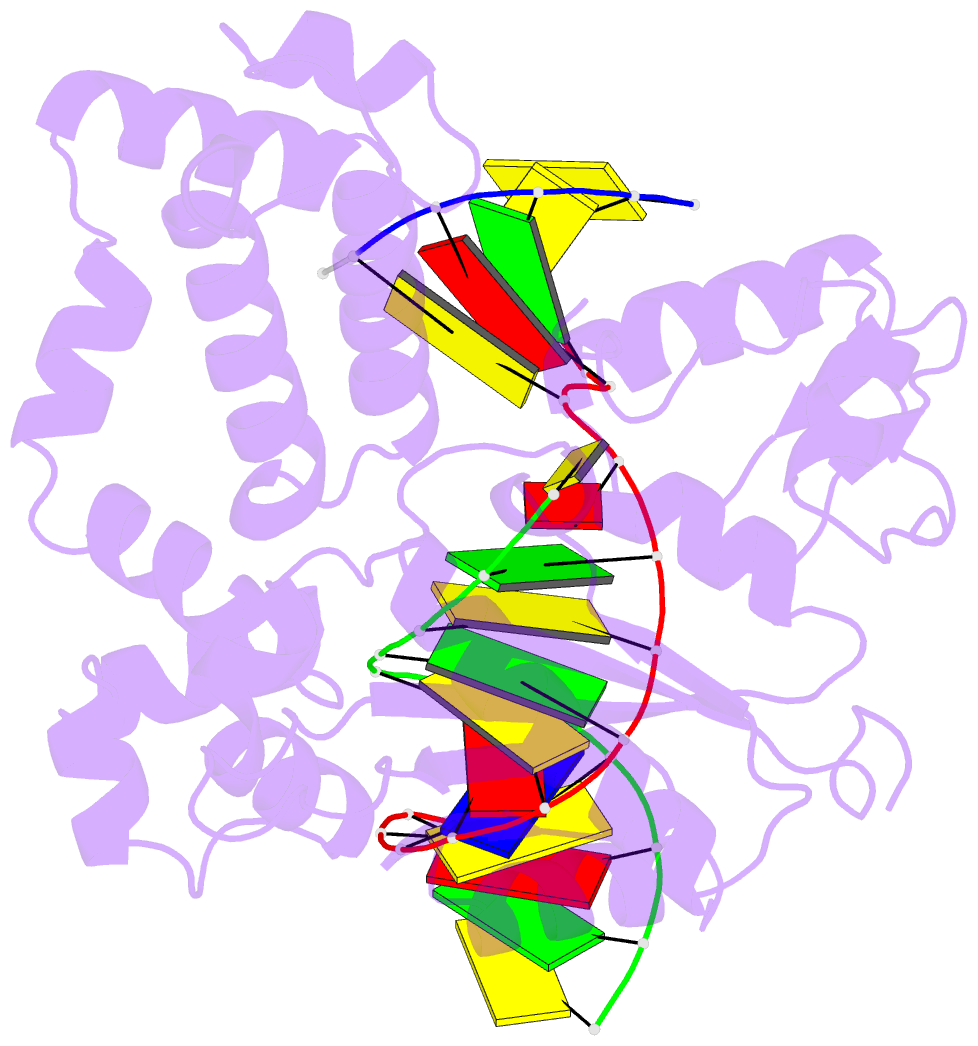
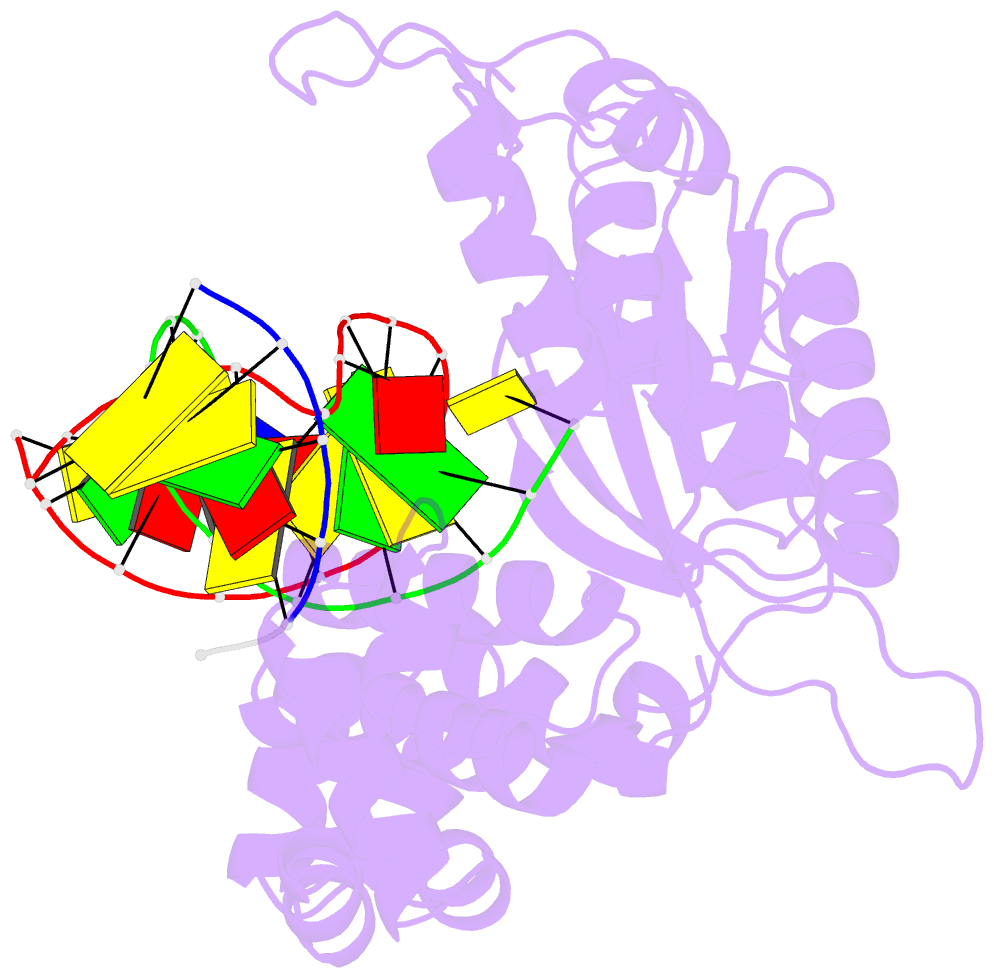
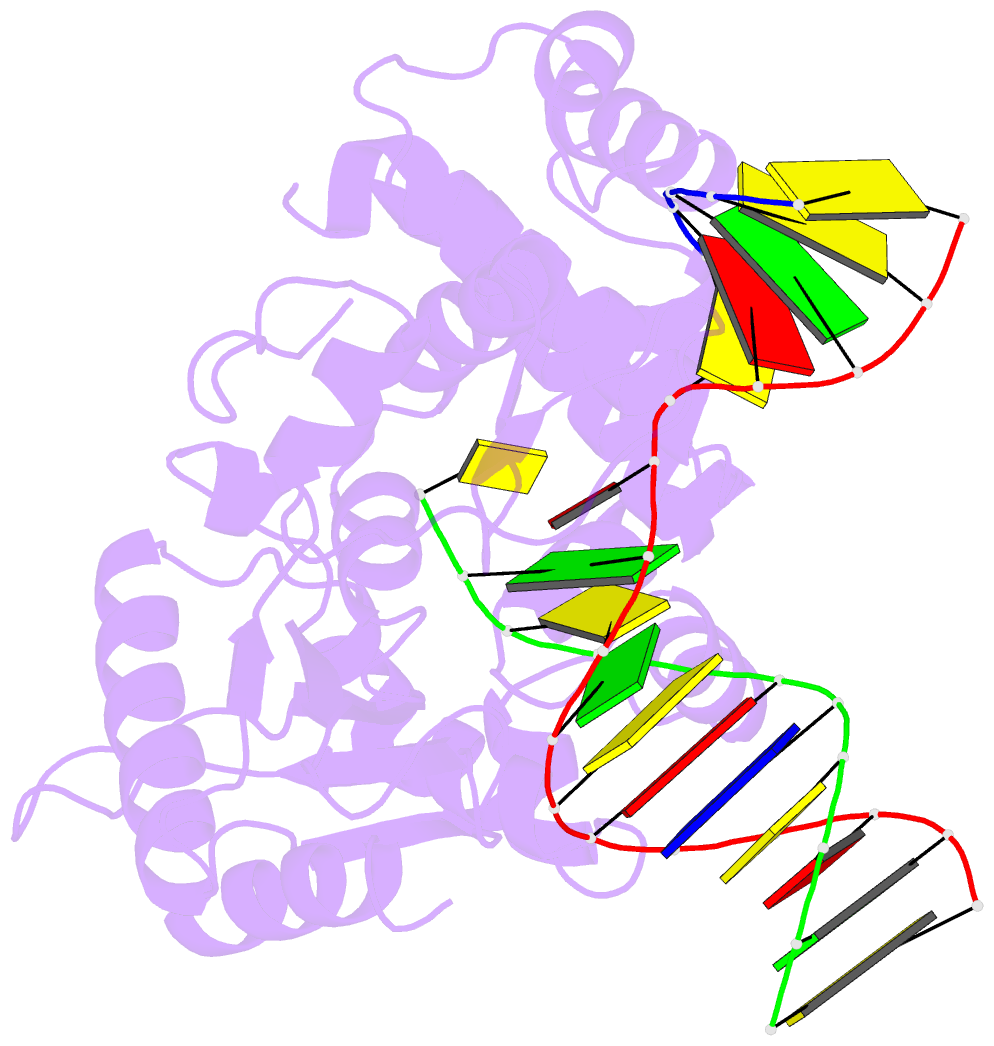
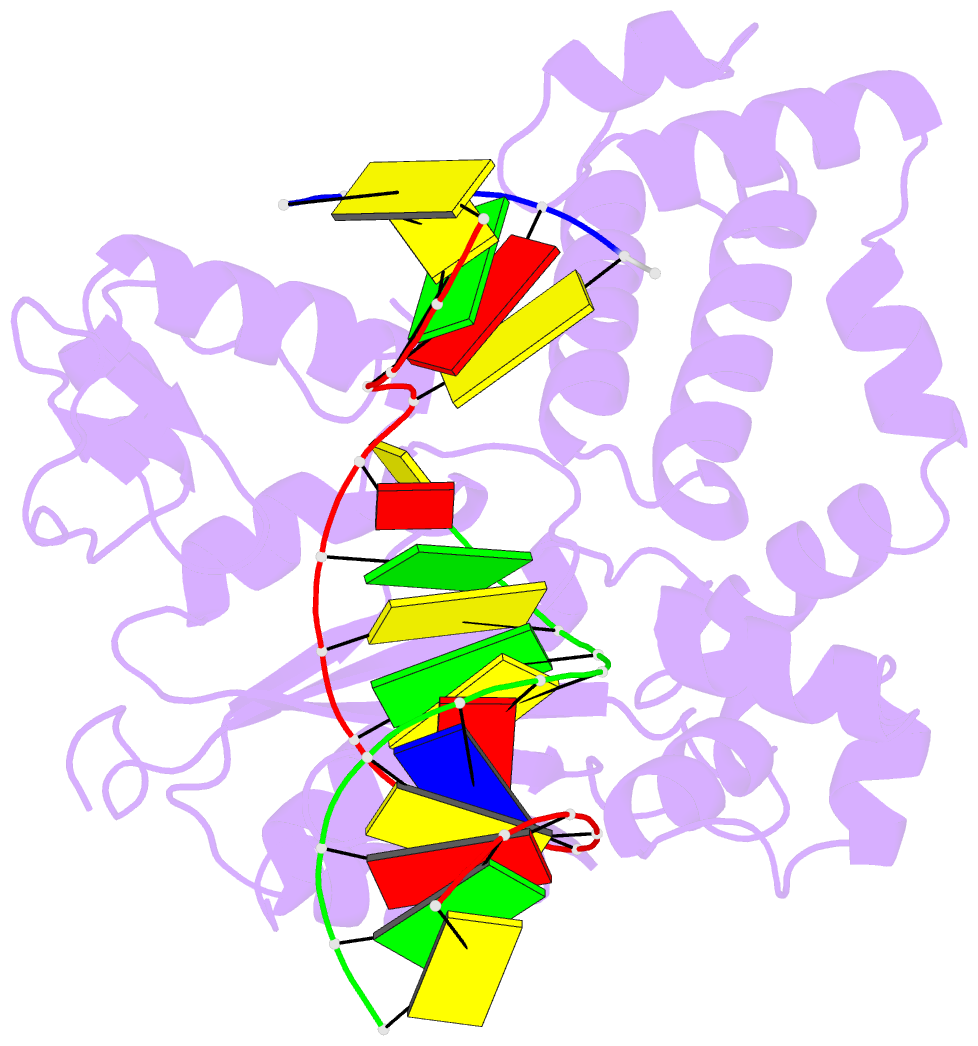
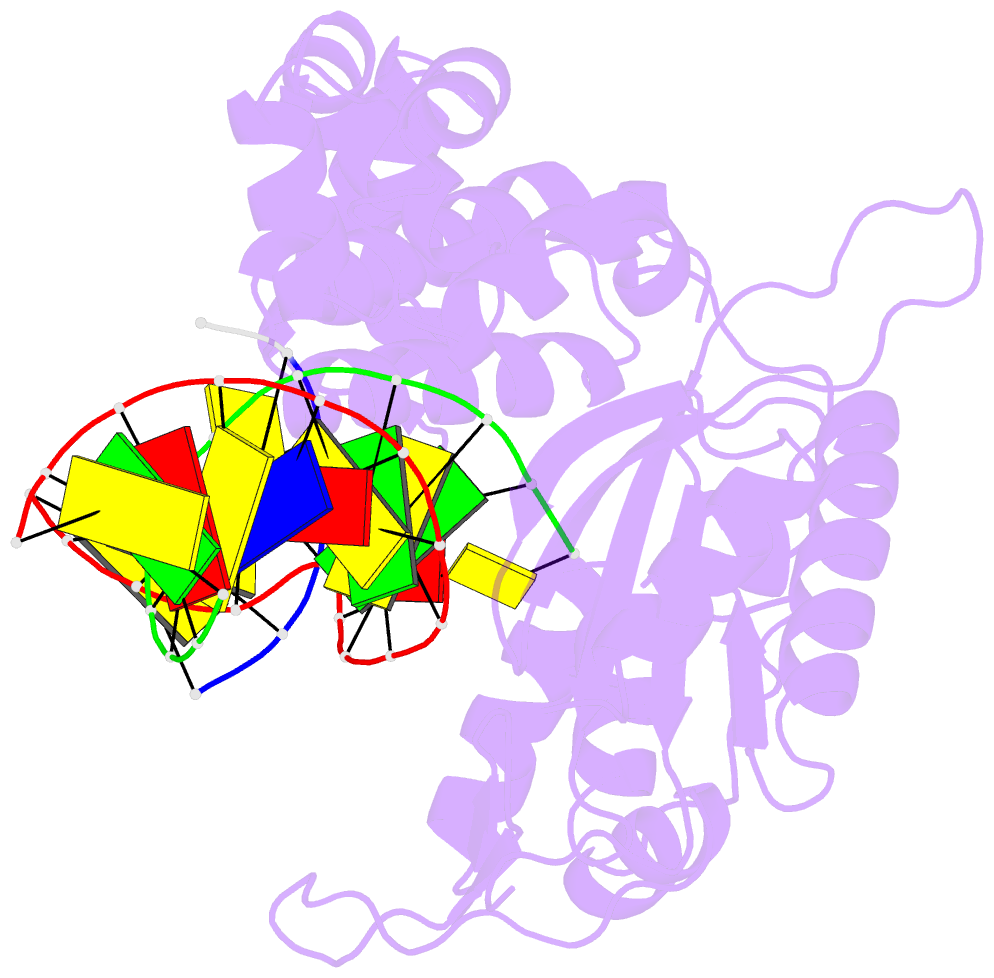
- Human DNA polymerase beta complexed with tetrahydrofuran (abasic site) containing DNA
- Reference
- Prasad R, Batra VK, Yang X-P, Krahn JM, Pedersen LC, Beard WA, Wilson SH (2005): "Structural insight into the DNA Polymerase beta deoxyribose phosphate lyase mechanism." DNA REPAIR, 4, 1347-1357. doi: 10.1016/j.dnarep.2005.08.009.
- Abstract
- A large number of biochemical and genetic studies have demonstrated the involvement of DNA polymerase beta (Pol beta) in mammalian base excision repair (BER). Pol beta participates in BER sub-pathways by contributing gap filling DNA synthesis and lyase removal of the 5'-deoxyribose phosphate (dRP) group from the cleaved abasic site. To better understand the mechanism of the dRP lyase reaction at an atomic level, we determined a crystal structure of Pol beta complexed with 5'-phosphorylated abasic sugar analogs in nicked DNA. This DNA ligand represents a potential BER intermediate. The crystal structure reveals that the dRP group is bound in a non-catalytic binding site. The catalytic nucleophile in the dRP lyase reaction, Lys72, and all other potential secondary nucleophiles, are too far away to participate in nucleophilic attack on the C1' of the sugar. An approximate model of the dRP group in the expected catalytic binding site suggests that a rotation of 120 degrees about the dRP 3'-phosphate is required to position the epsilon-amino Lys72 close to the dRP C1'. This model also suggests that several other side chains are in position to facilitate the beta-elimination reaction. From results of mutational analysis of key residues in the dRP lyase active site, it appears that the substrate dRP can be stabilized in the observed non-catalytic binding conformation, hindering dRP lyase activity.