Summary information and primary citation
- PDB-id
- 2pfj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (3.1 Å)
- Summary
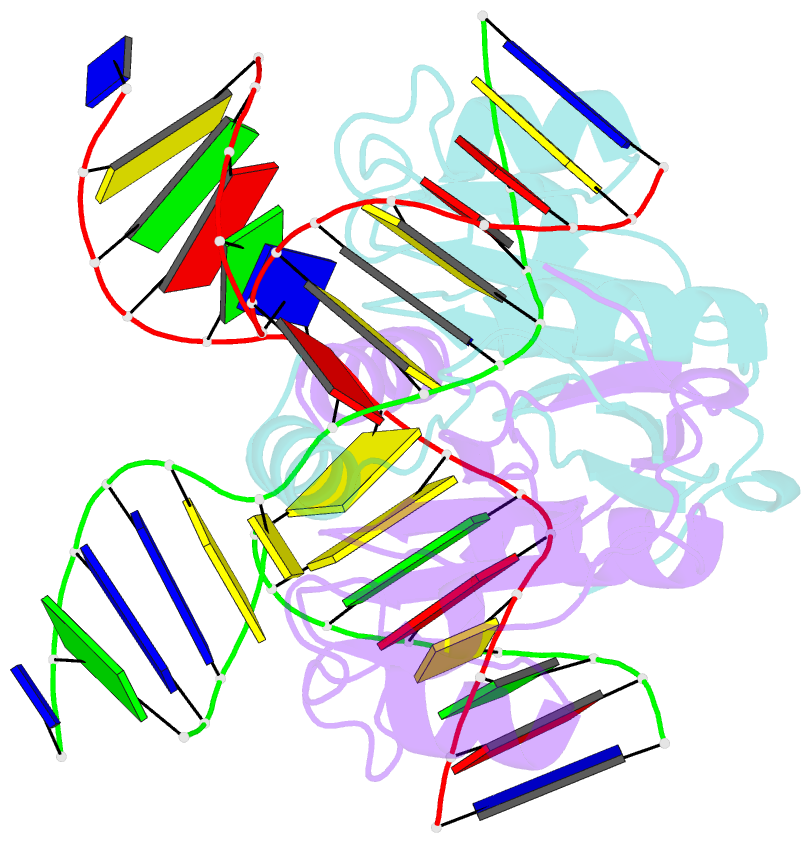
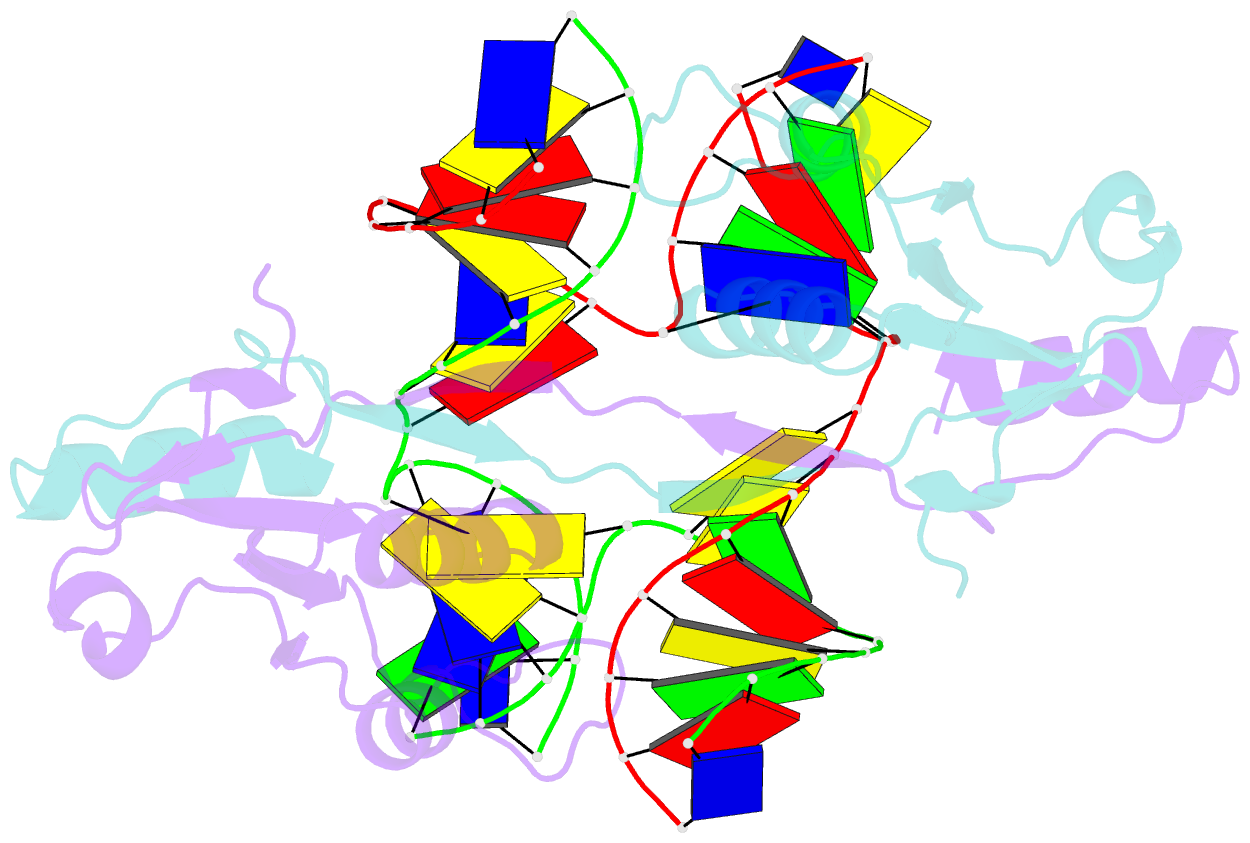
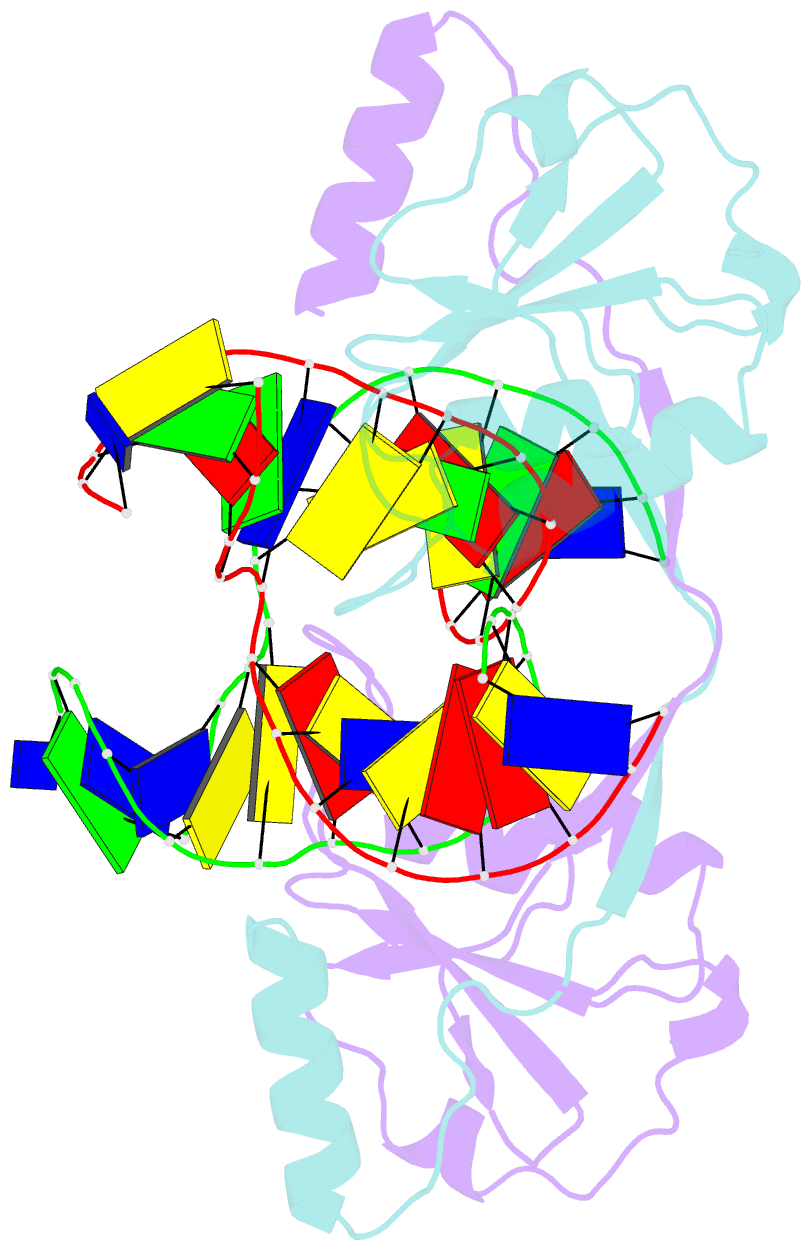
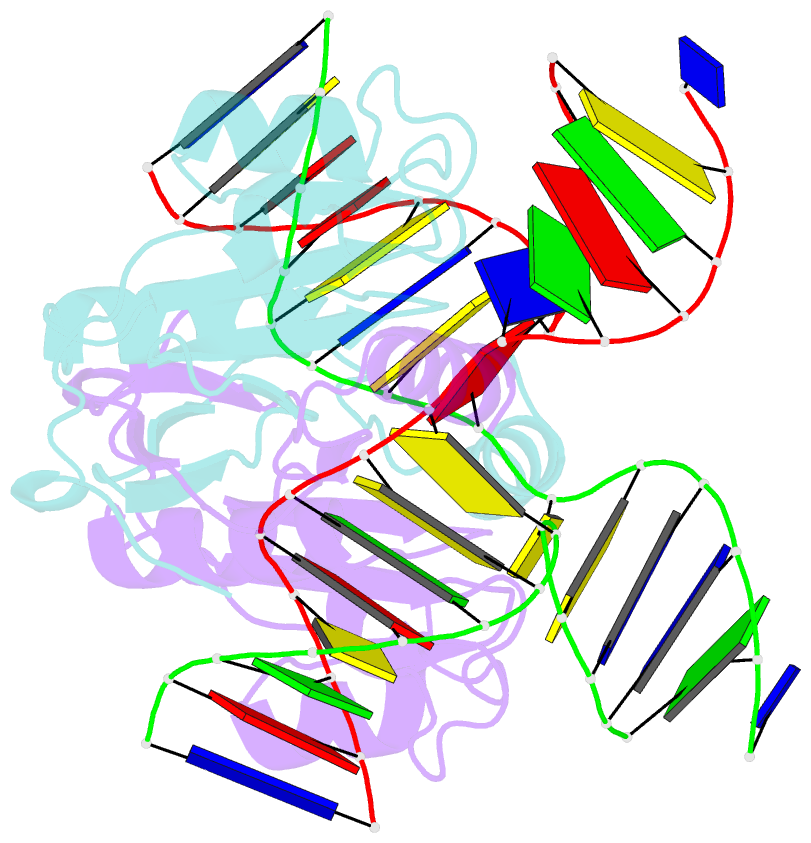
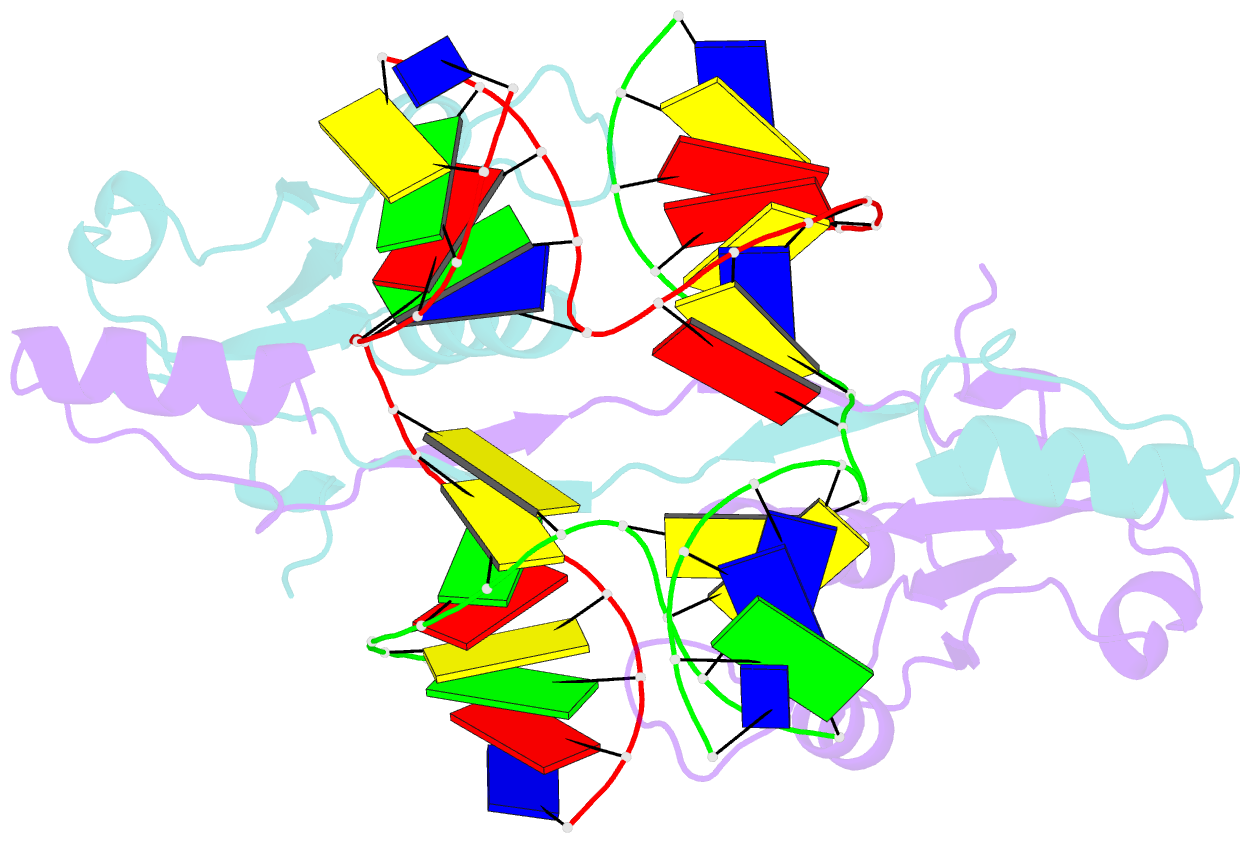
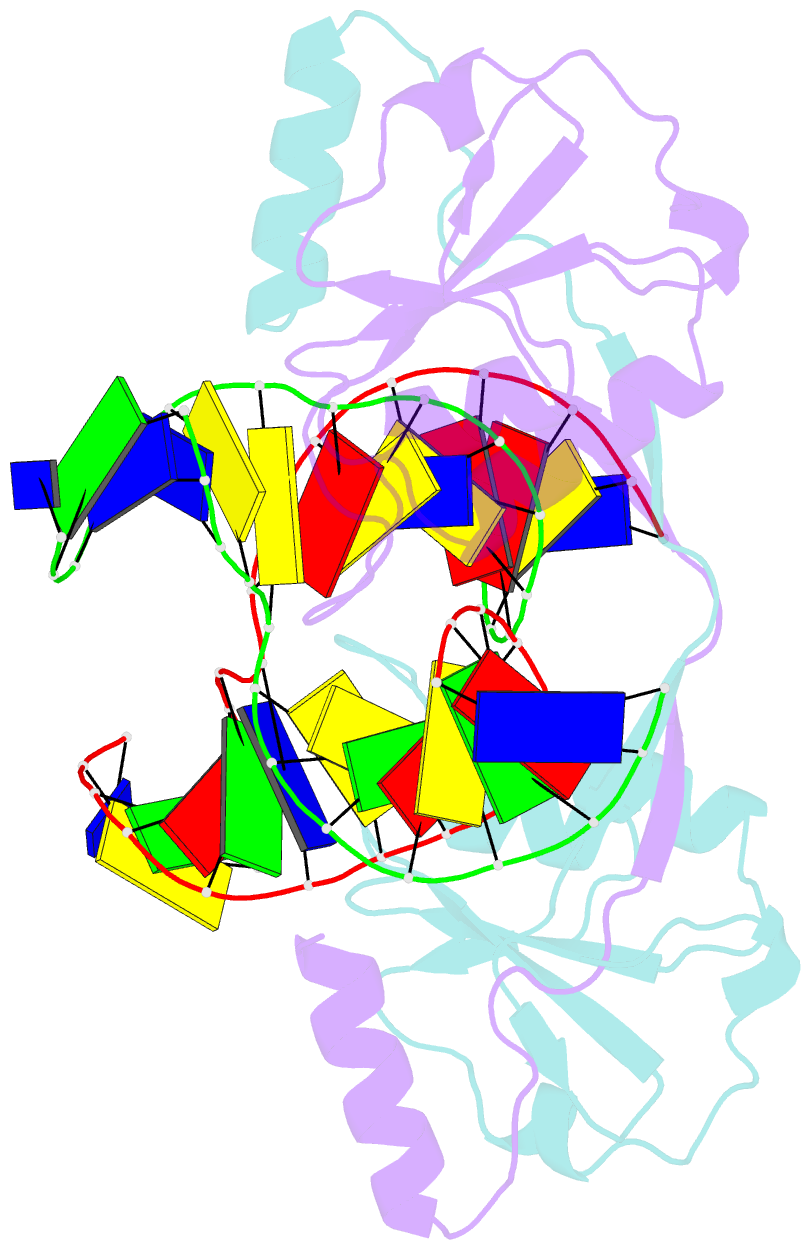
- Crystal structure of t7 endo i resolvase in complex with a holliday junction
- Reference
- Hadden JM, Declais AC, Carr SB, Lilley DM, Phillips SE (2007): "The structural basis of Holliday junction resolution by T7 endonuclease I." Nature, 449, 621-624. doi: 10.1038/nature06158.
- Abstract
- The four-way (Holliday) DNA junction is the central intermediate in homologous recombination, a ubiquitous process that is important in DNA repair and generation of genetic diversity. The penultimate stage of recombination requires resolution of the DNA junction into nicked-duplex species by the action of a junction-resolving enzyme, examples of which have been identified in a wide variety of organisms. These enzymes are nucleases that are highly selective for the structure of branched DNA. The mechanism of this selectivity has, however, been unclear in the absence of structural data. Here we present the crystal structure of the junction-resolving enzyme phage T7 endonuclease I in complex with a synthetic four-way DNA junction. Although the enzyme is structure-selective, significant induced fit occurs in the interaction, with changes in the structure of both the protein and the junction. The dimeric enzyme presents two binding channels that contact the backbones of the junction's helical arms over seven nucleotides. These interactions effectively measure the relative orientations and positions of the arms of the junction, thereby ensuring that binding is selective for branched DNA that can achieve this geometry.