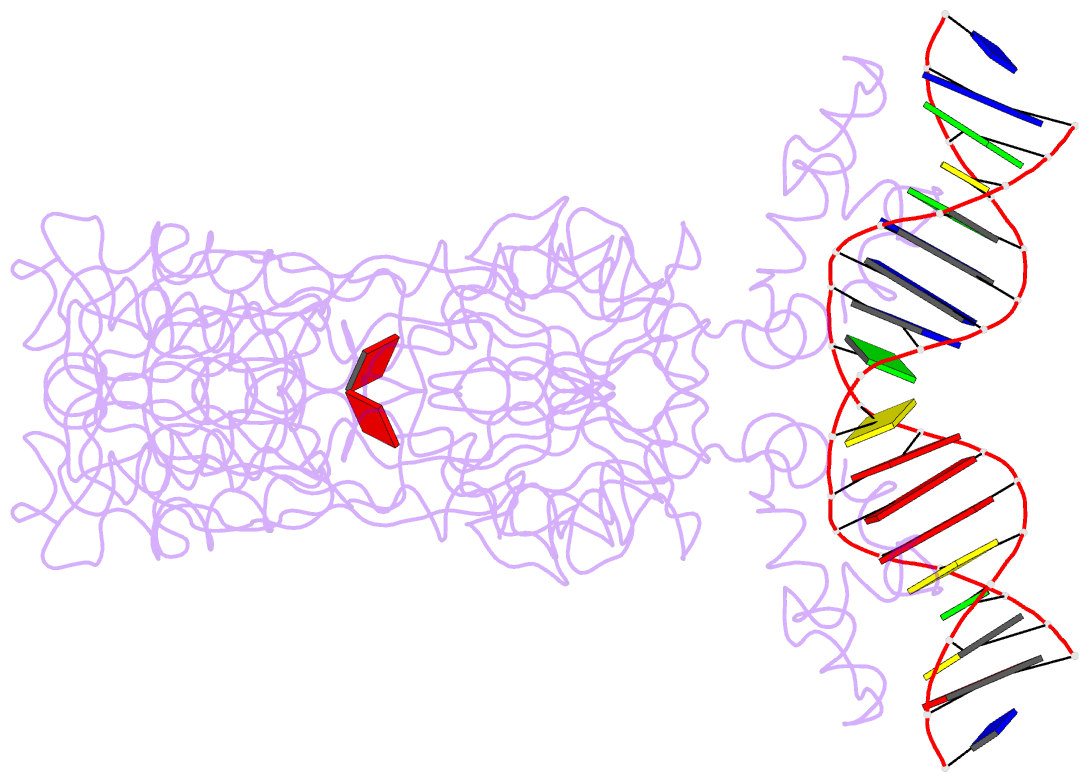
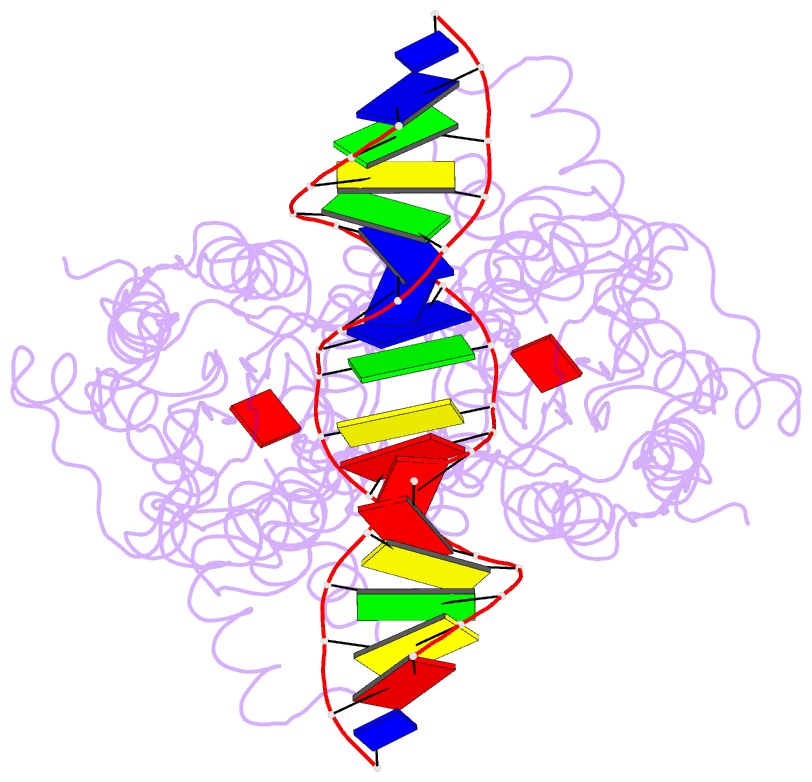
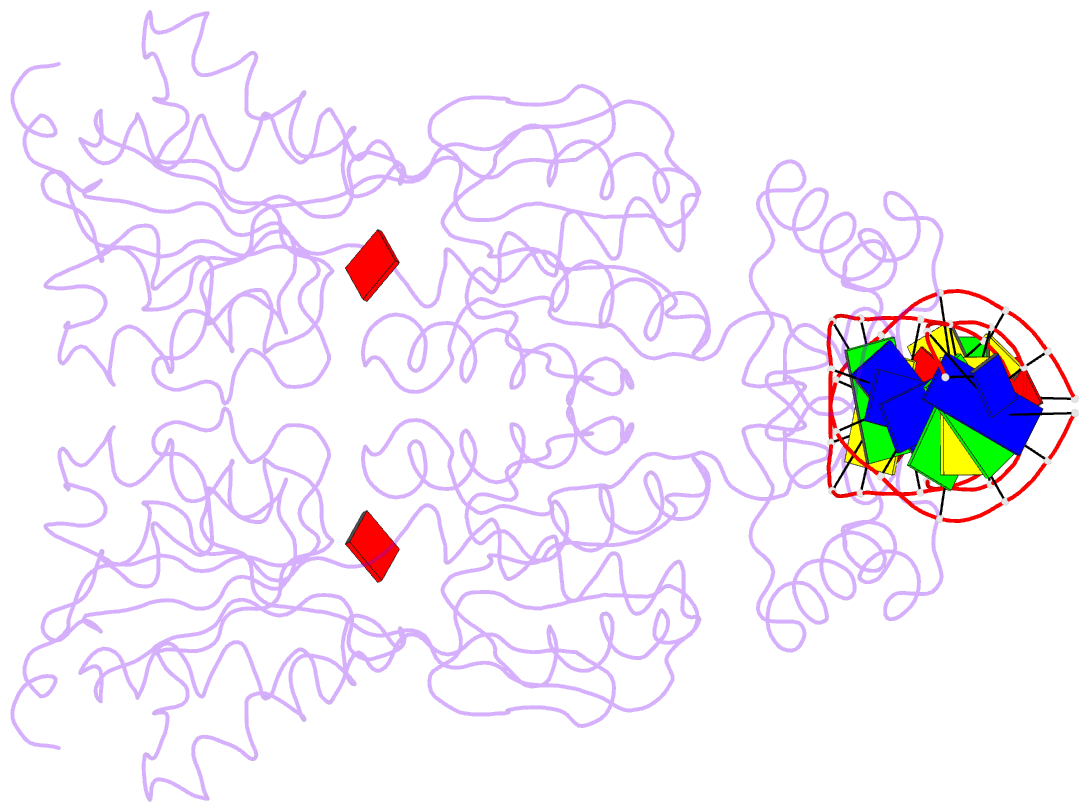
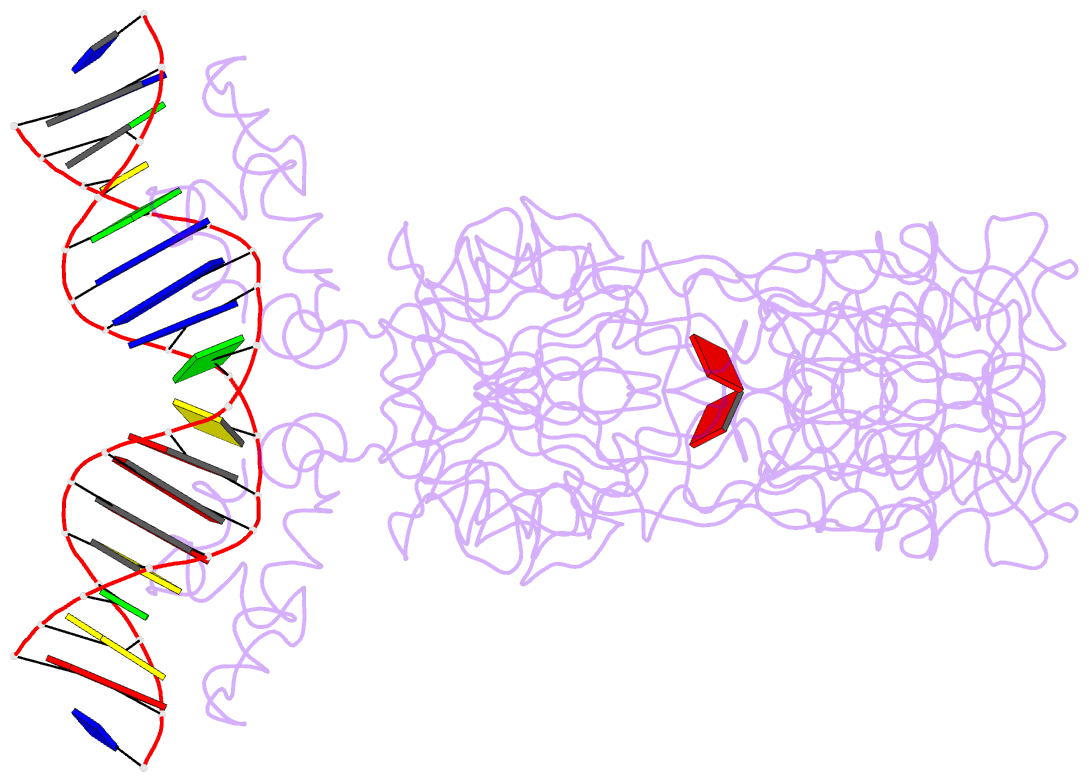
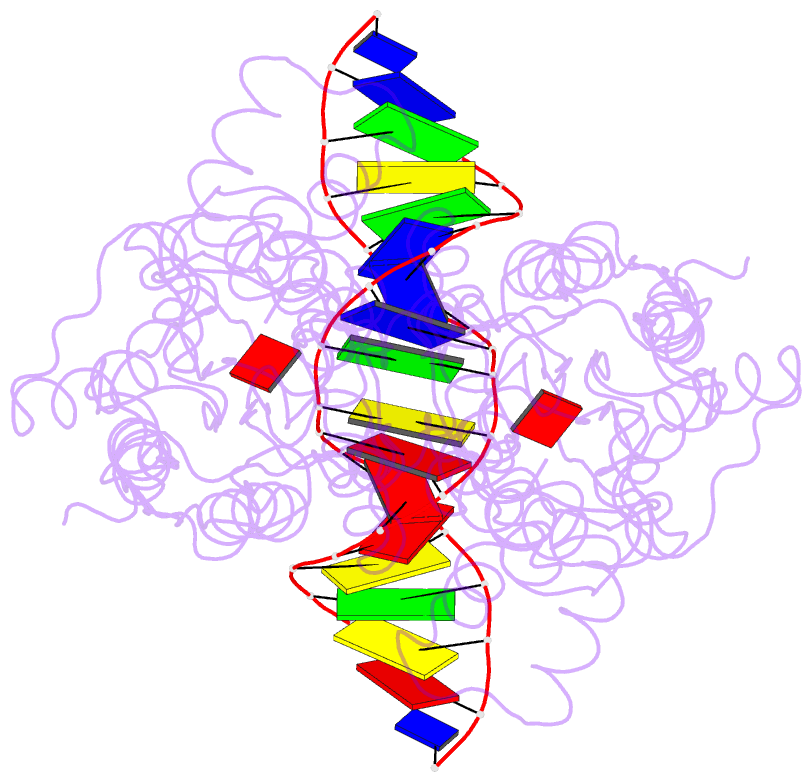
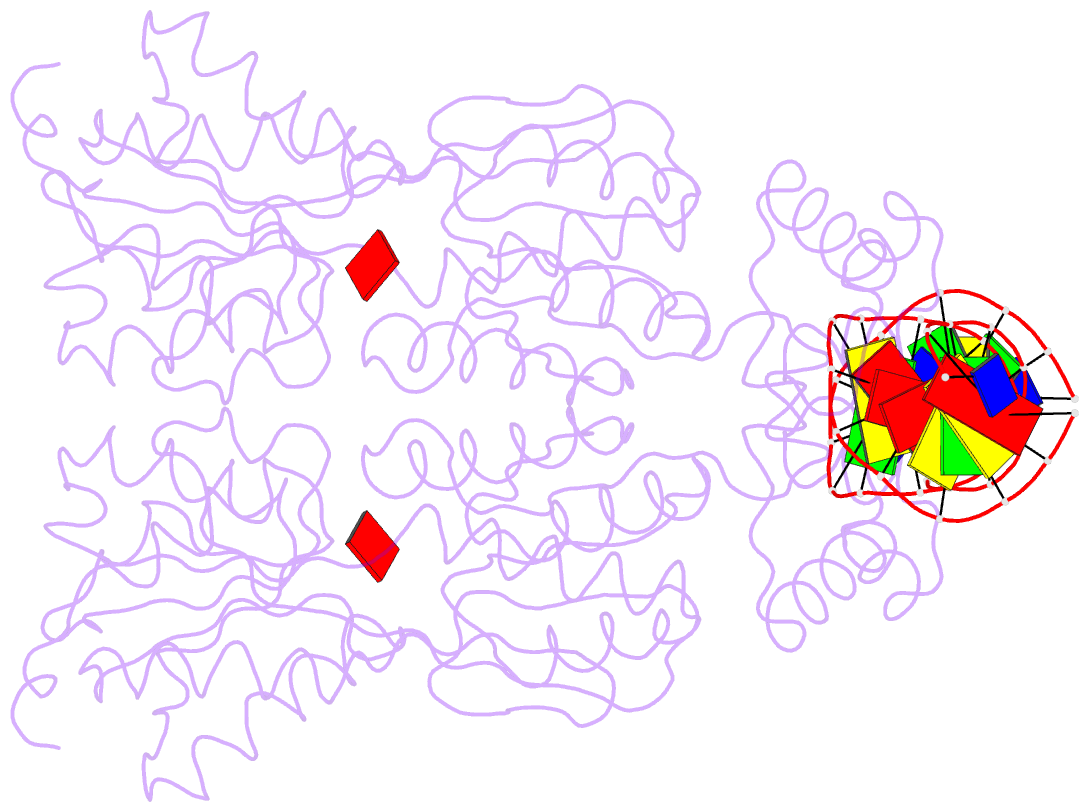
Summary information and primary citation
- PDB-id
- 2pue; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.7 Å)
- Summary
- Crystal structure of the laci family member, purr, bound to DNA: minor groove binding by alpha helices
- Reference
- Lu F, Schumacher MA, Arvidson DN, Haldimann A, Wanner BL, Zalkin H, Brennan RG (1998): "Structure-based redesign of corepressor specificity of the Escherichia coli purine repressor by substitution of residue 190." Biochemistry, 37, 971-982. doi: 10.1021/bi971942s.
- Abstract
- Guanine or hypoxanthine, physiological corepressors of the Escherichia coli purine repressor (PurR), promote formation of the ternary PurR-corepressor-operator DNA complex that functions to repress pur operon gene expression. Structure-based predictions on the importance of Arg190 in determining 6-oxopurine specificity and corepressor binding affinity were tested by mutagenesis, analysis of in vivo function, and in vitro corepressor binding measurements. Replacements of Arg190 with Ala or Gln resulted in functional repressors in which binding of guanine and hypoxanthine was retained but specificity was relaxed to permit binding of adenine. X-ray structures were determined for ternary complexes of mutant repressors with purines (adenine, guanine, hypoxanthine, and 6-methylpurine) and operator DNA. These structures indicate that R190A binds guanine, hypoxanthine, and adenine with nearly equal, albeit reduced, affinity in large part because of a newly made compensatory hydrogen bond between the rotated hydroxyl side chain of Ser124 and the exocyclic 6 positions of the purines. Through direct and water-mediated contacts, the R190Q protein binds adenine with a nearly 75-fold higher affinity than the wild type repressor while maintaining wild type affinity for guanine and hypoxanthine. The results establish at the atomic level the basis for the critical role of Arg190 in the recognition of the exocyclic 6 position of its purine corepressors and the successful redesign of corepressor specificity.