Summary information and primary citation
- PDB-id
- 2pyo; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- structural protein-DNA
- Method
- X-ray (2.43 Å)
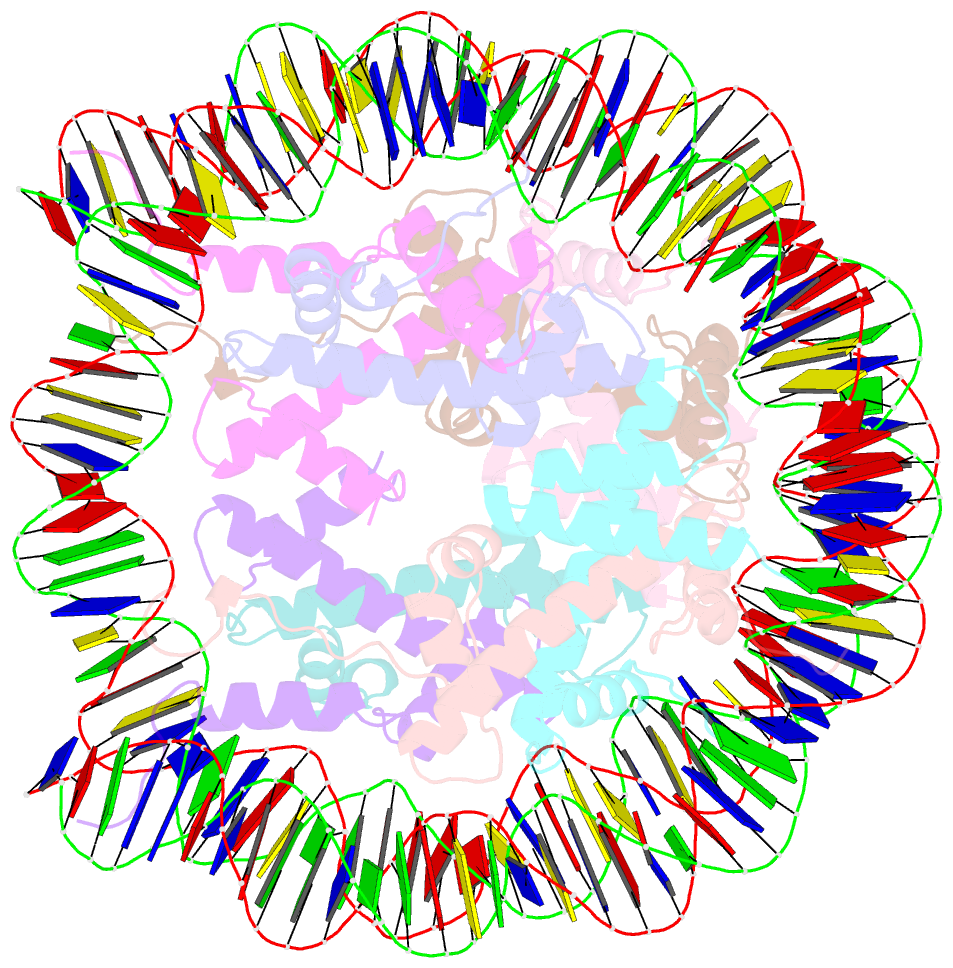
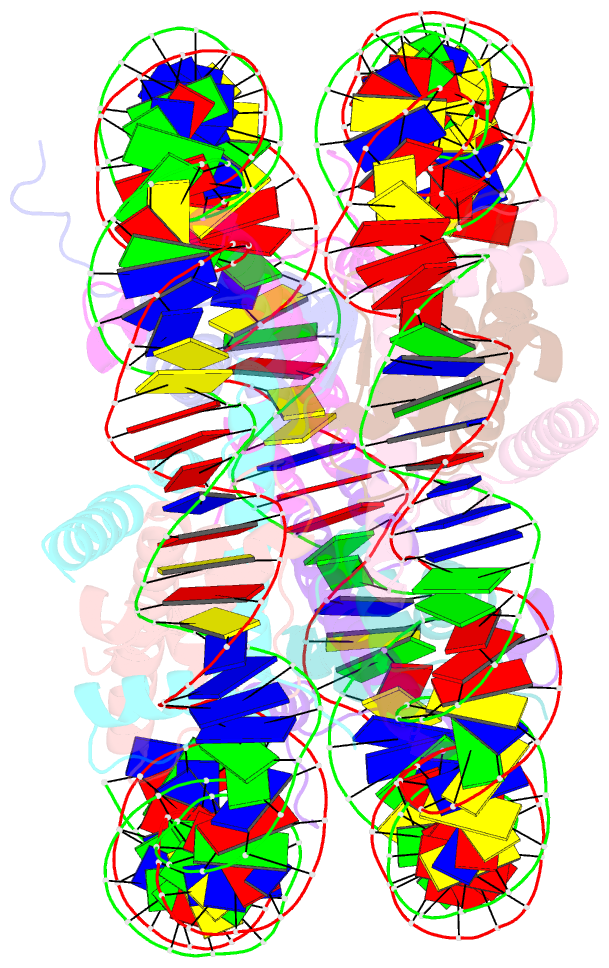
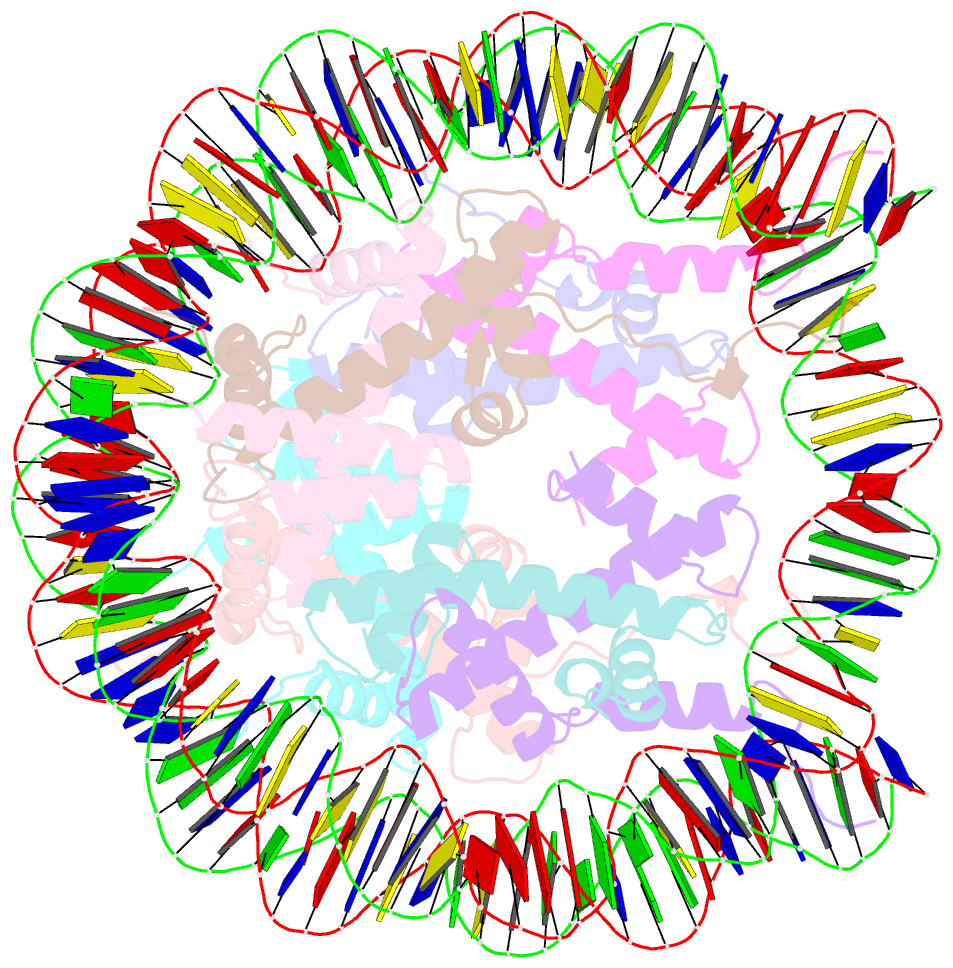
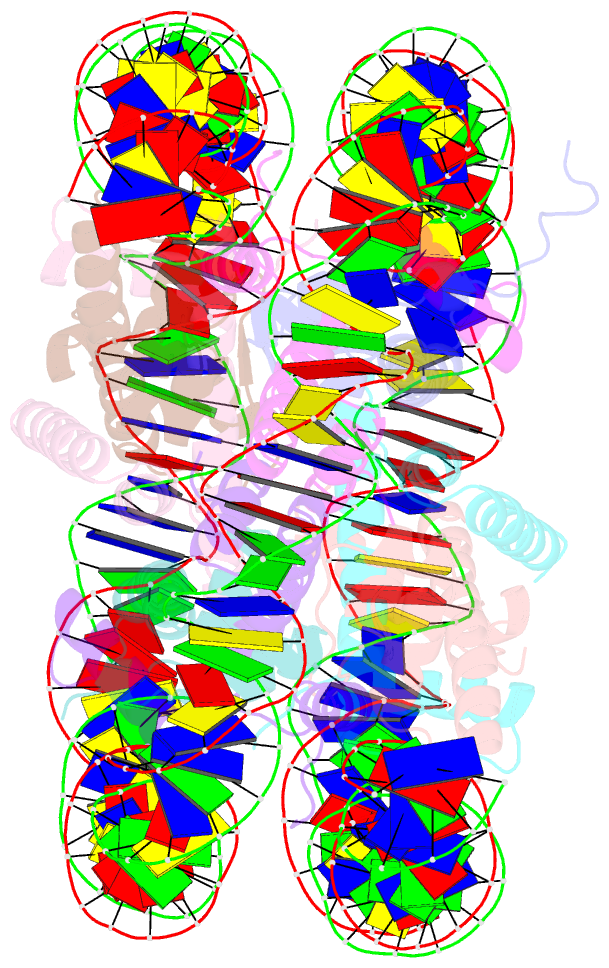
- Summary
- Drosophila nucleosome core
- Reference
- Clapier CR, Chakravarthy S, Petosa C, Fernandez-Tornero C, Luger K, Muller CW (2007): "Structure of the Drosophila nucleosome core particle highlights evolutionary constraints on the H2A-H2B histone dimer." Proteins, 71, 1-7. doi: 10.1002/prot.21720.
- Abstract
- We determined the 2.45 A crystal structure of the nucleosome core particle from Drosophila melanogaster and compared it to that of Xenopus laevis bound to the identical 147 base-pair DNA fragment derived from human alpha-satellite DNA. Differences between the two structures primarily reflect 16 amino acid substitutions between species, 15 of which are in histones H2A and H2B. Four of these involve histone tail residues, resulting in subtly altered protein-DNA interactions that exemplify the structural plasticity of these tails. Of the 12 substitutions occurring within the histone core regions, five involve small, solvent-exposed residues not involved in intraparticle interactions. The remaining seven involve buried hydrophobic residues, and appear to have coevolved so as to preserve the volume of side chains within the H2A hydrophobic core and H2A-H2B dimer interface. Thus, apart from variations in the histone tails, amino acid substitutions that differentiate Drosophila from Xenopus histones occur in mutually compensatory combinations. This highlights the tight evolutionary constraints exerted on histones since the vertebrate and invertebrate lineages diverged.