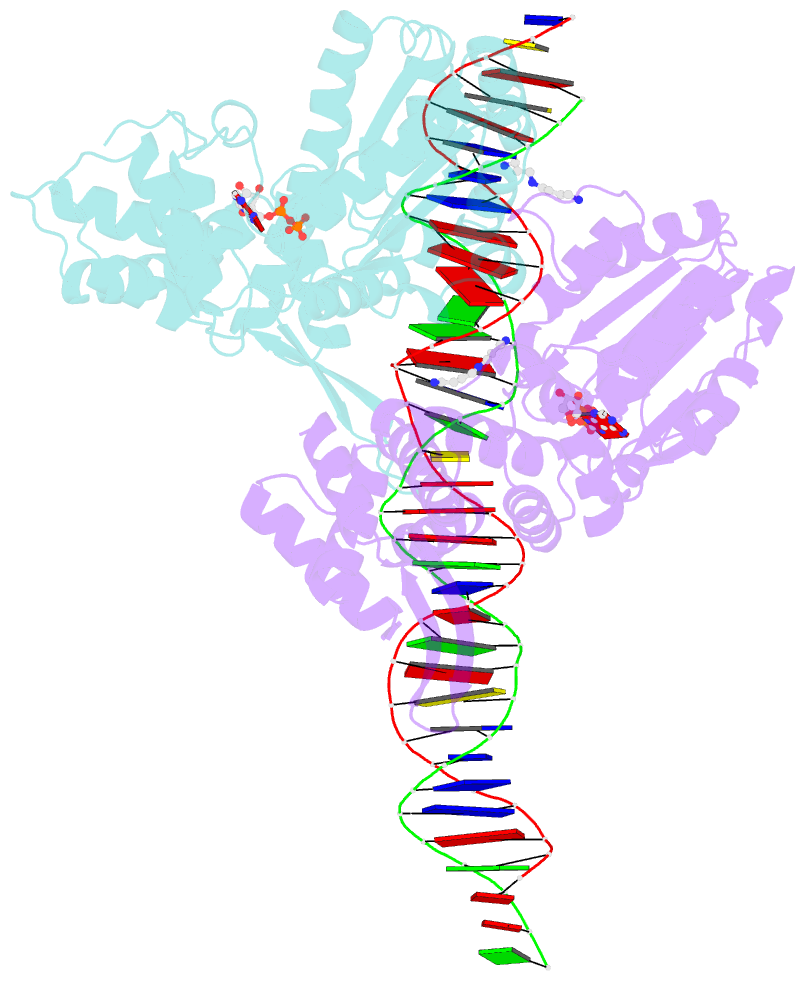
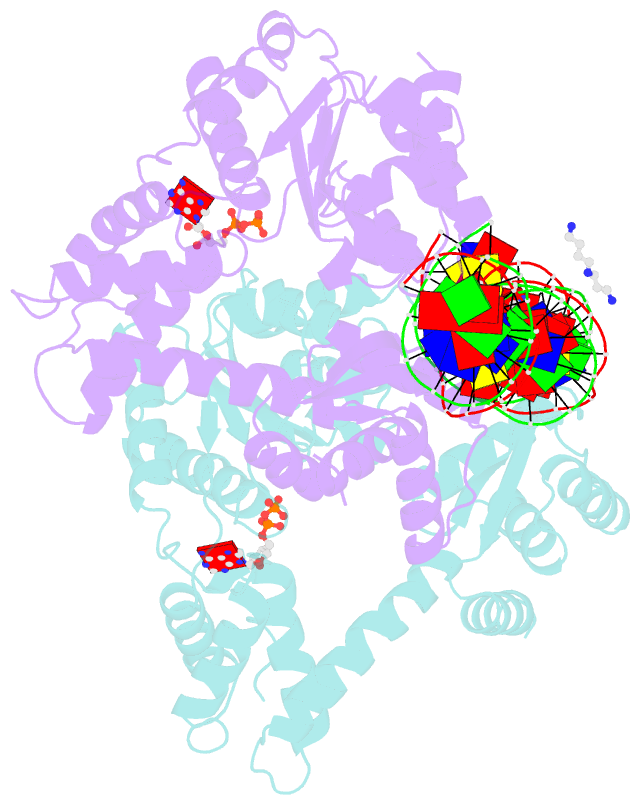
Summary information and primary citation
- PDB-id
- 2qby; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication-DNA
- Method
- X-ray (3.35 Å)
- Summary
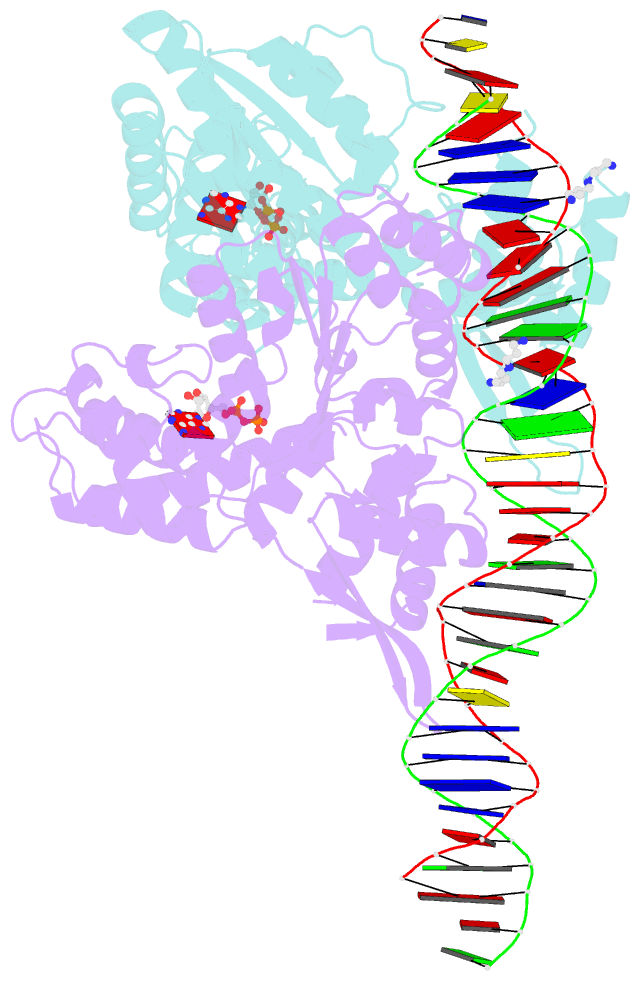
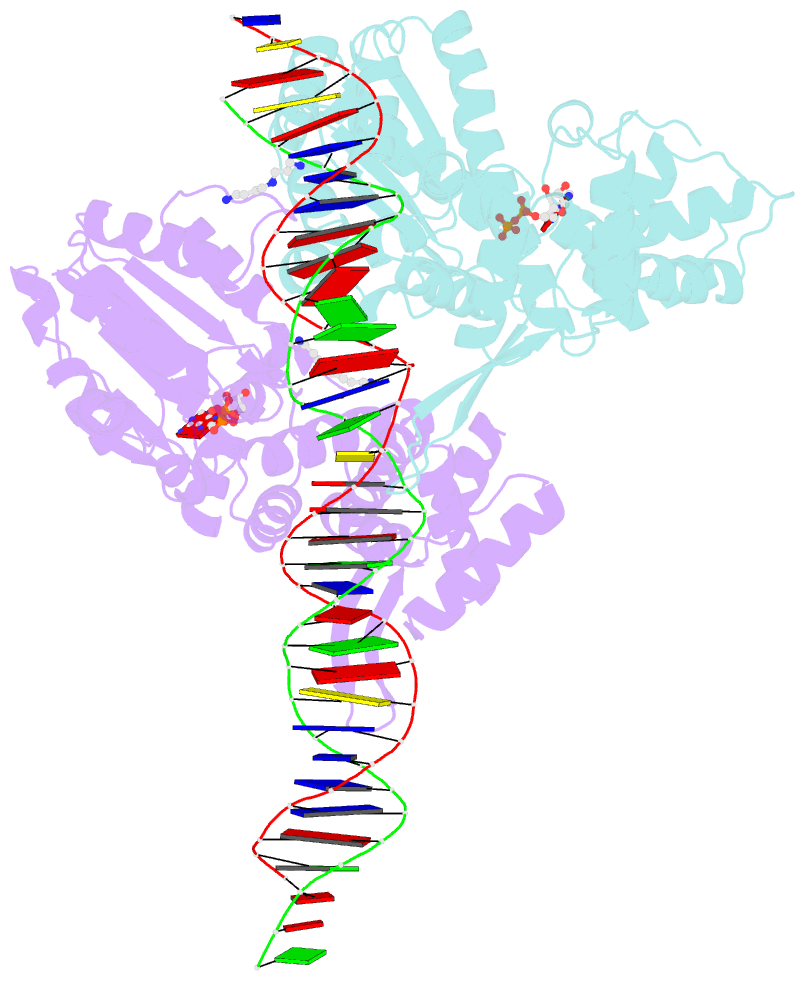
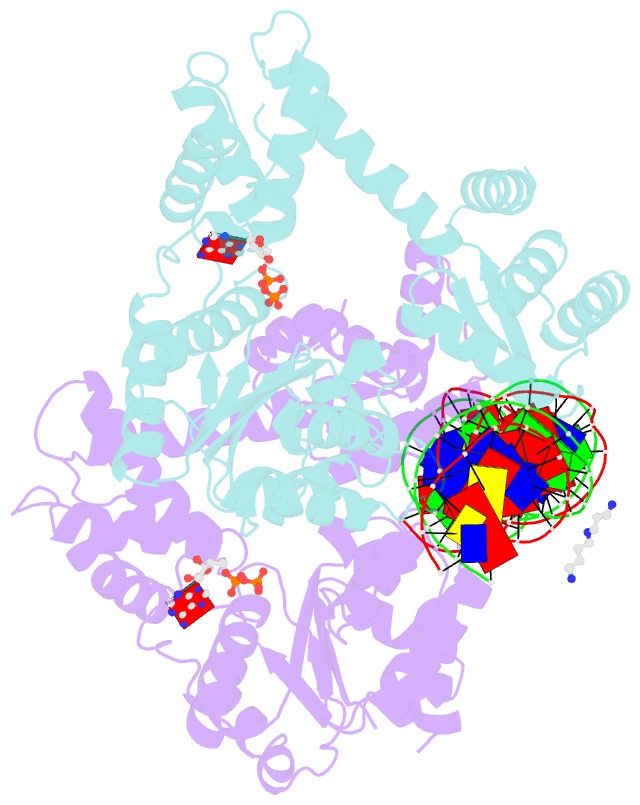
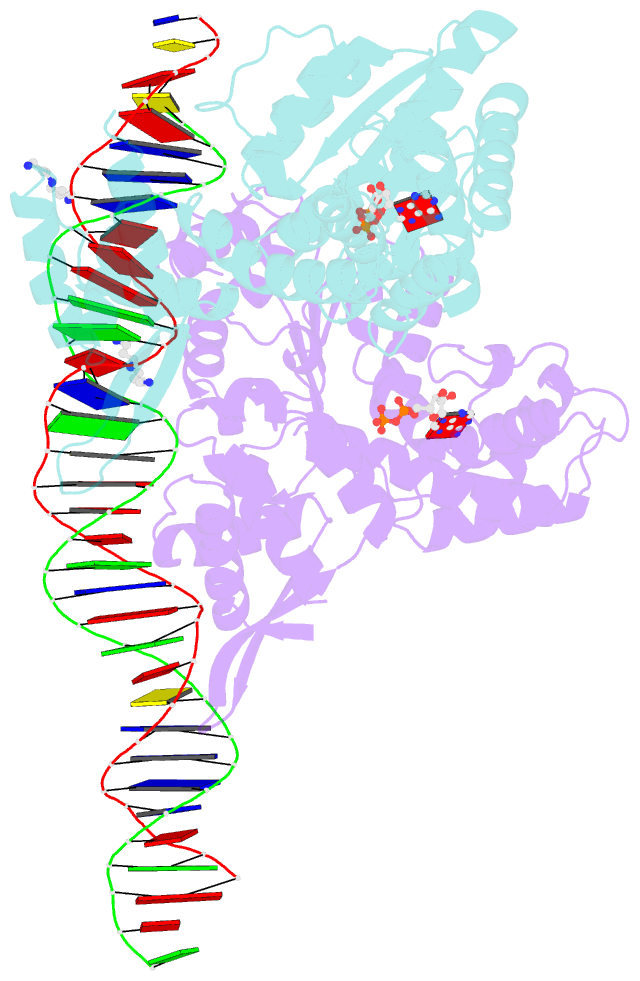
- Crystal structure of a heterodimer of cdc6-orc1 initiators bound to origin DNA (from s. solfataricus)
- Reference
- Dueber EL, Corn JE, Bell SD, Berger JM (2007): "Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex." Science, 317, 1210-1213. doi: 10.1126/science.1143690.
- Abstract
- The faithful duplication of genetic material depends on essential DNA replication initiation factors. Cellular initiators form higher-order assemblies on replication origins, using adenosine triphosphate (ATP) to locally remodel duplex DNA and facilitate proper loading of synthetic replisomal components. To better understand initiator function, we determined the 3.4 angstrom-resolution structure of an archaeal Cdc6/Orc1 heterodimer bound to origin DNA. The structure demonstrates that, in addition to conventional DNA binding elements, initiators use their AAA+ ATPase domains to recognize origin DNA. Together these interactions establish the polarity of initiator assembly on the origin and induce substantial distortions into origin DNA strands. Biochemical and comparative analyses indicate that AAA+/DNA contacts observed in the structure are dynamic and evolutionarily conserved, suggesting that the complex forms a core component of the basal initiation machinery.