Summary information and primary citation
- PDB-id
- 2qfj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription repressor-DNA
- Method
- X-ray (2.1 Å)
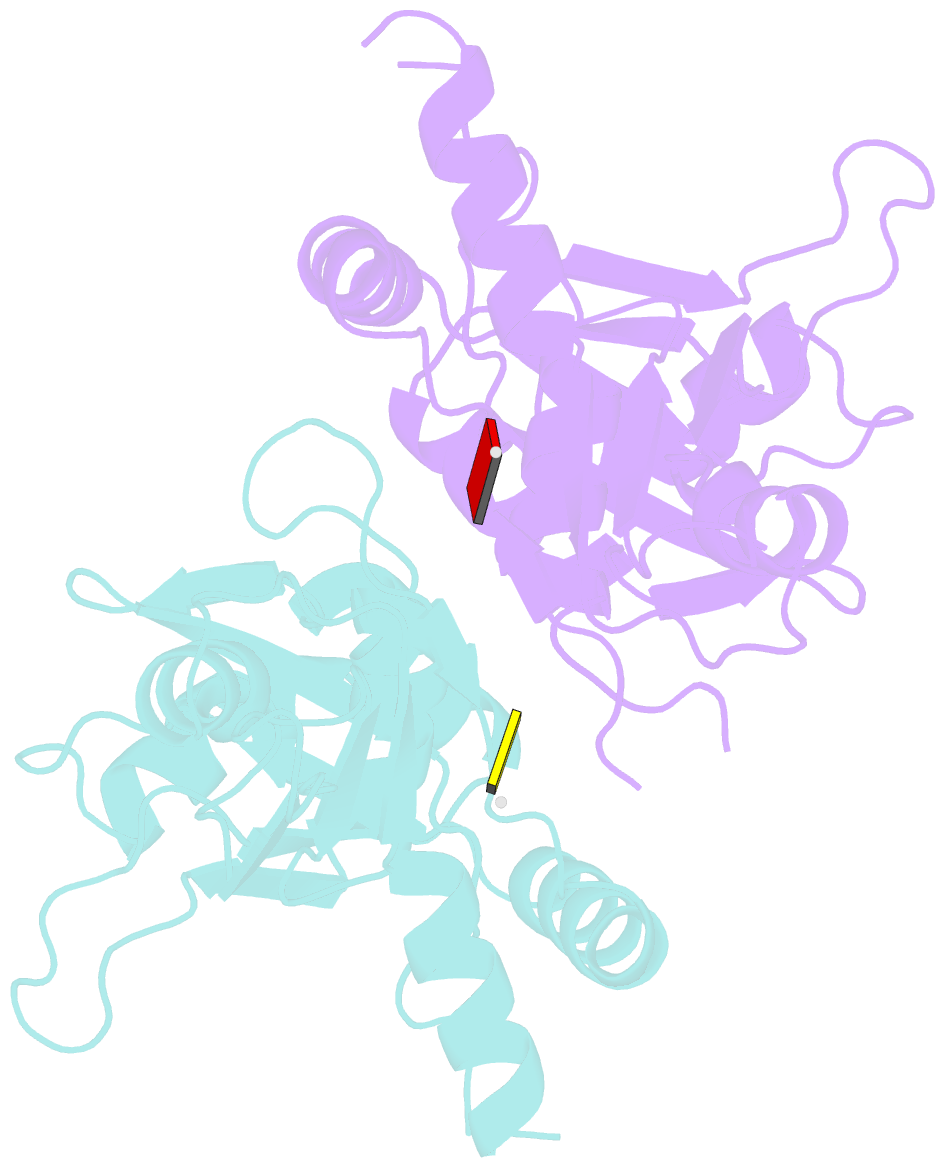
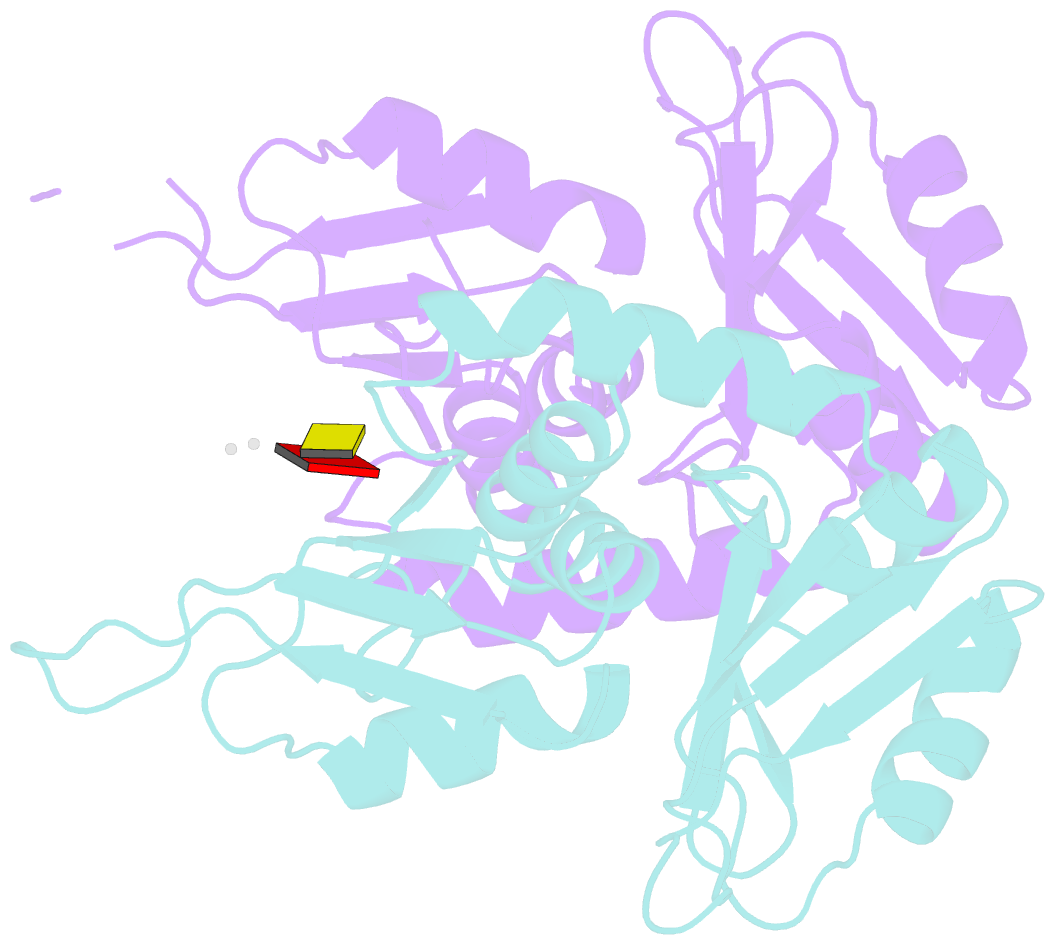
- Summary
- Crystal structure of first two rrm domains of fir bound to ssDNA from a portion of fuse
- Reference
- Crichlow GV, Zhou H, Hsiao H-H, Frederick KB, Debrosse M, Yang Y, Folta-Stogniew EJ, Chung H-J, Fan C, De La Cruz EM, Levens D, Lolis E, Braddock D (2007): "Dimerization of FIR upon FUSE DNA binding suggests a mechanism of c-myc inhibition." EMBO J., 27, 277-289. doi: 10.1038/sj.emboj.7601936.
- Abstract
- c-myc is essential for cell homeostasis and growth but lethal if improperly regulated. Transcription of this oncogene is governed by the counterbalancing forces of two proteins on TFIIH--the FUSE binding protein (FBP) and the FBP-interacting repressor (FIR). FBP and FIR recognize single-stranded DNA upstream of the P1 promoter, known as FUSE, and influence transcription by oppositely regulating TFIIH at the promoter site. Size exclusion chromatography coupled with light scattering reveals that an FIR dimer binds one molecule of single-stranded DNA. The crystal structure confirms that FIR binds FUSE as a dimer, and only the N-terminal RRM domain participates in nucleic acid recognition. Site-directed mutations of conserved residues in the first RRM domain reduce FIR's affinity for FUSE, while analogous mutations in the second RRM domain either destabilize the protein or have no effect on DNA binding. Oppositely oriented DNA on parallel binding sites of the FIR dimer results in spooling of a single strand of bound DNA, and suggests a mechanism for c-myc transcriptional control.
Cartoon-block schematics in six views