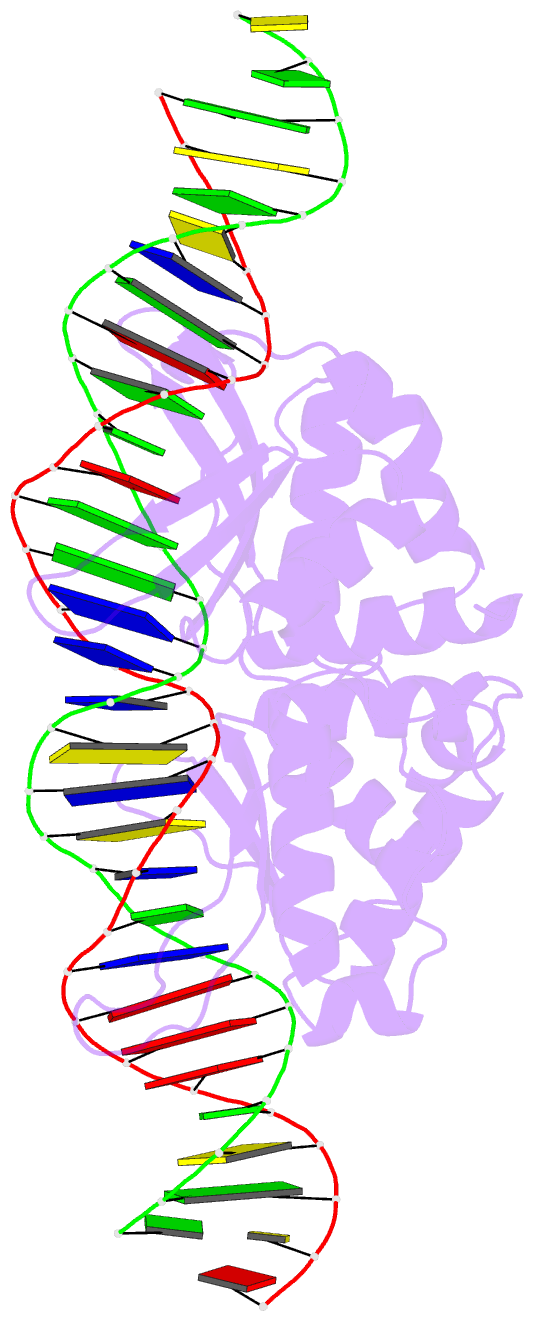
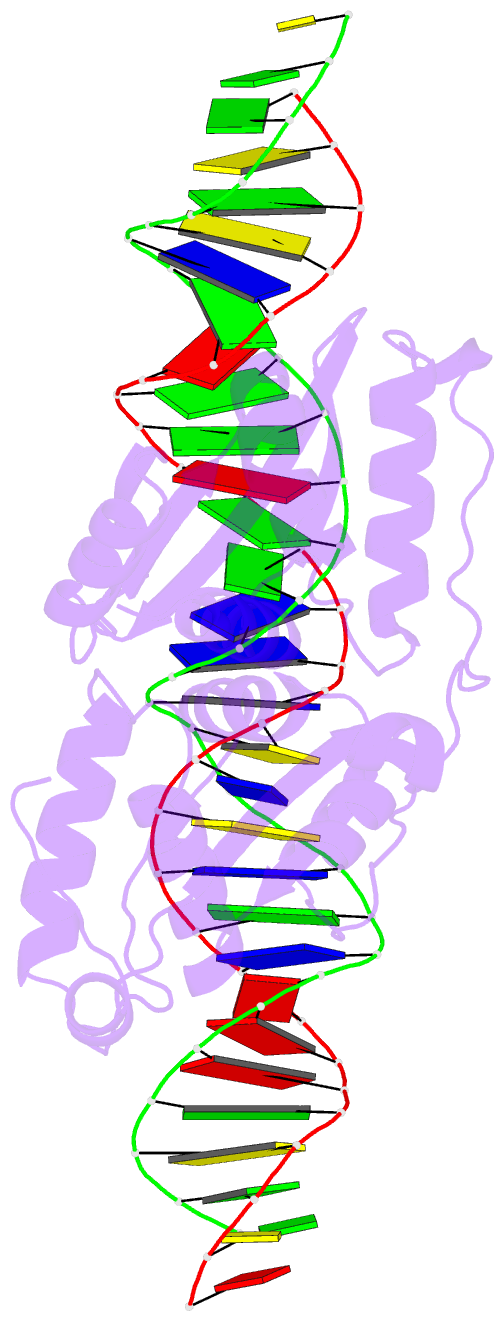
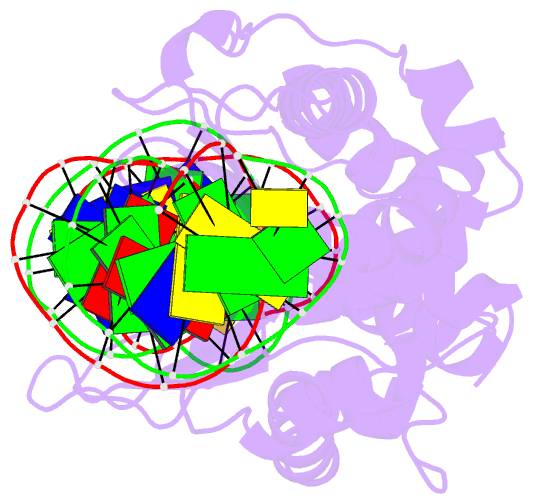
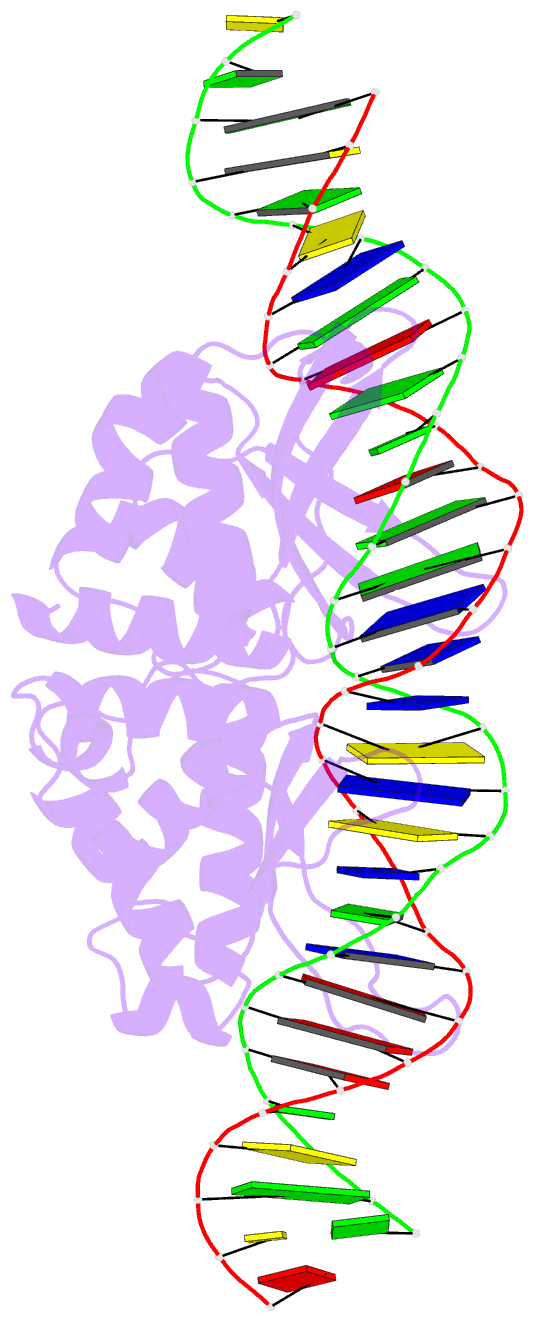
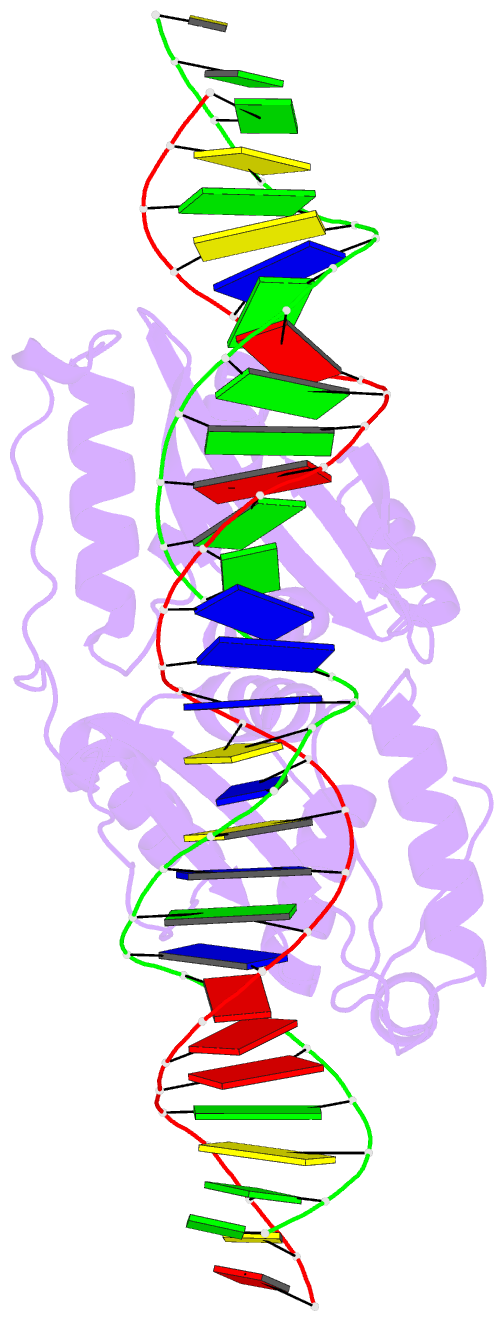
Summary information and primary citation
- PDB-id
- 2qoj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.4 Å)
- Summary
- Coevolution of a homing endonuclease and its host target sequence
- Reference
- Scalley-Kim M, McConnell-Smith A, Stoddard BL (2007): "Coevolution of a homing endonuclease and its host target sequence." J.Mol.Biol., 372, 1305-1319. doi: 10.1016/j.jmb.2007.07.052.
- Abstract
- We have determined the specificity profile of the homing endonuclease I-AniI and compared it to the conservation of its host gene. Homing endonucleases are encoded within intervening sequences such as group I introns. They initiate the transfer of such elements by cleaving cognate alleles lacking the intron, leading to their transfer via homologous recombination. Each structural homing endonuclease family has arrived at an appropriate balance of specificity and fidelity that avoids toxicity while maximizing target recognition and invasiveness. I-AniI recognizes a strongly conserved target sequence in a host gene encoding apocytochrome B and has fine-tuned its specificity to correlate with wobble versus nonwobble positions across that sequence and to the amount of degeneracy inherent in individual codons. The physiological target site in the host gene is not the optimal substrate for recognition and cleavage: at least one target variant identified during a screen is bound more tightly and cleaved more rapidly. This is a result of the periodic cycle of intron homing, which at any time can present nonoptimal combinations of endonuclease specificity and insertion site sequences in a biological host.