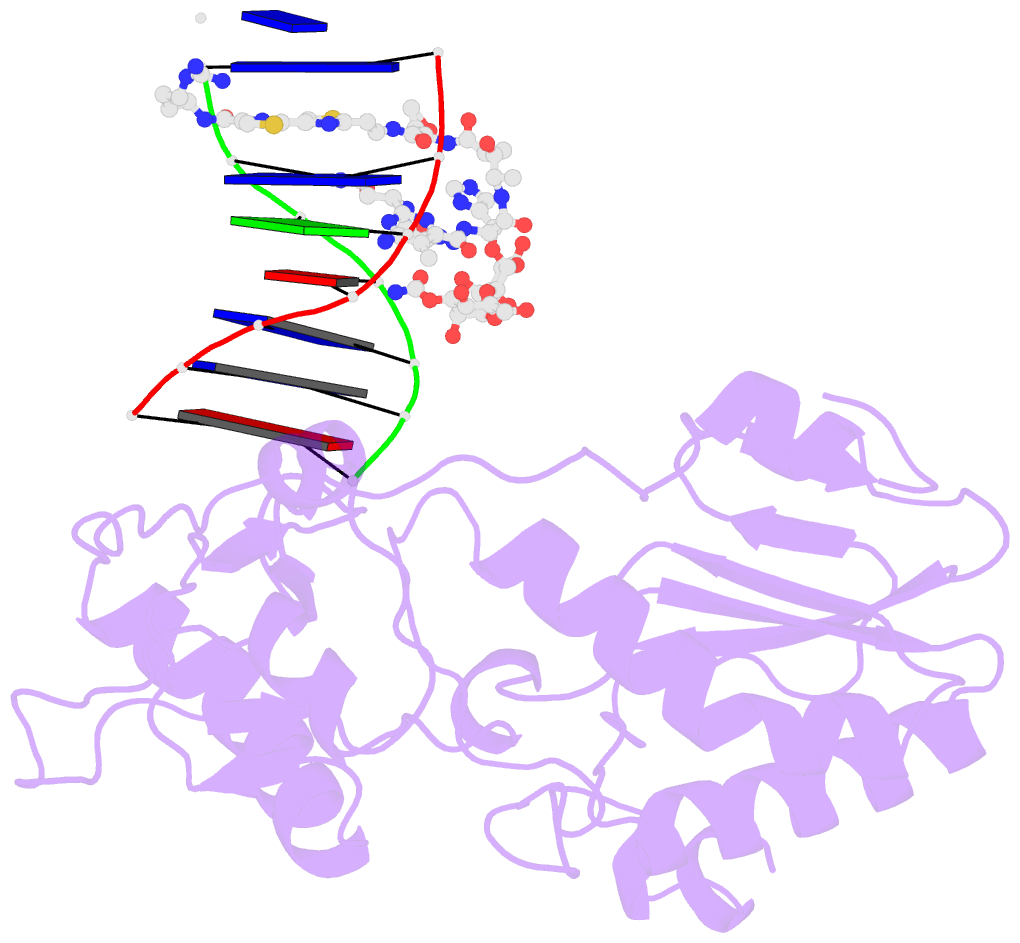
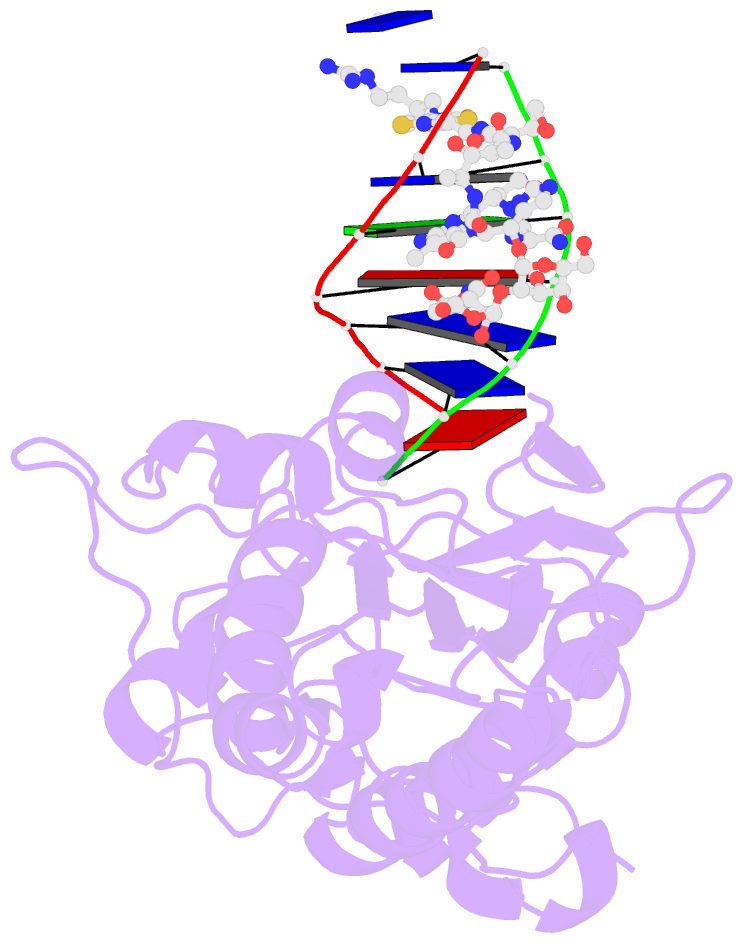
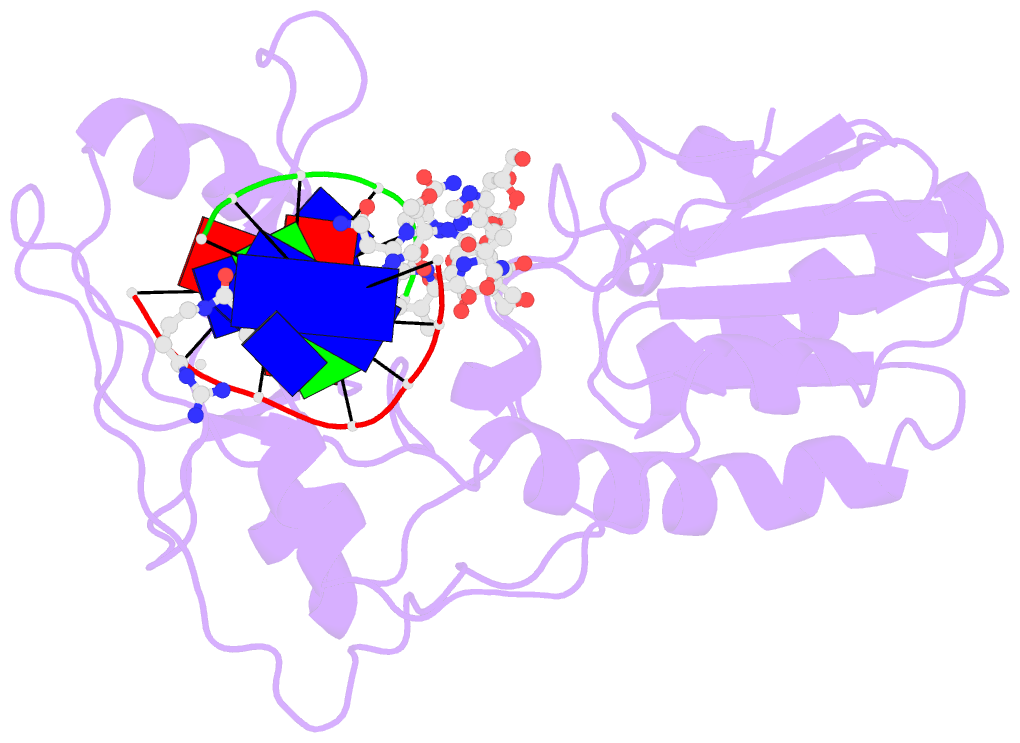
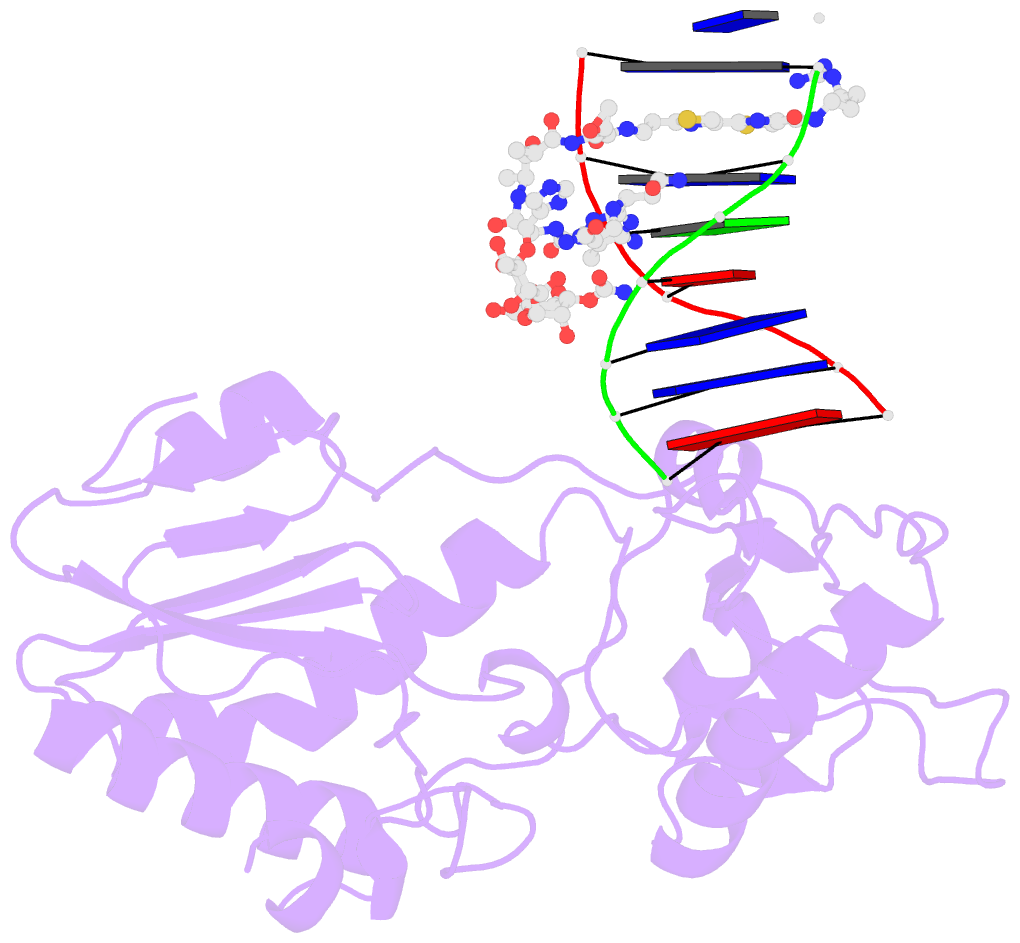
Summary information and primary citation
- PDB-id
- 2r2s; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.8 Å)
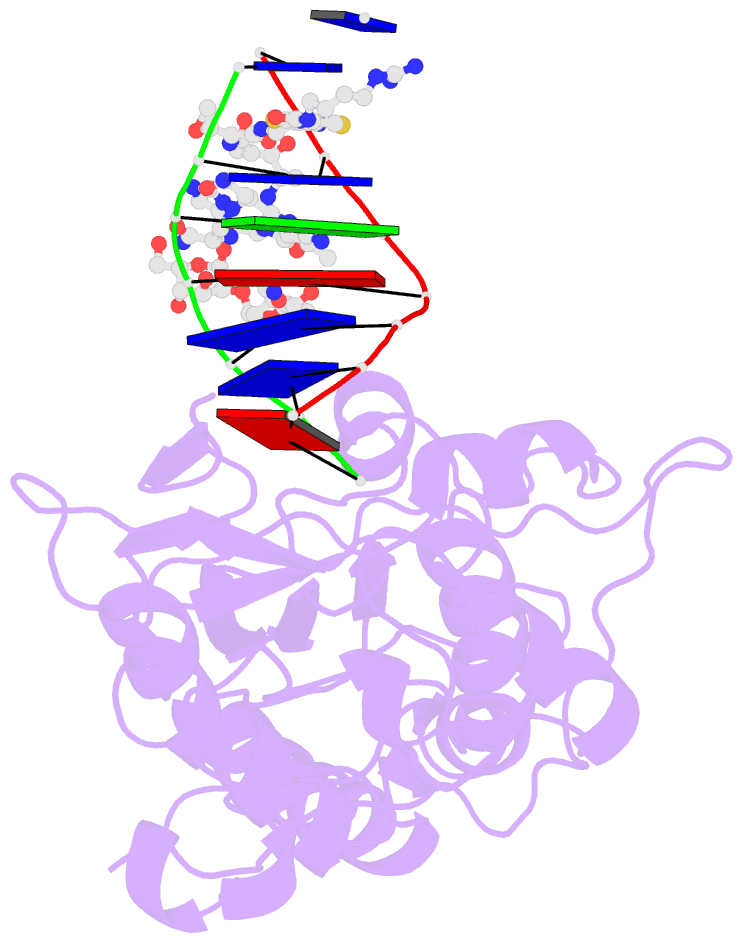
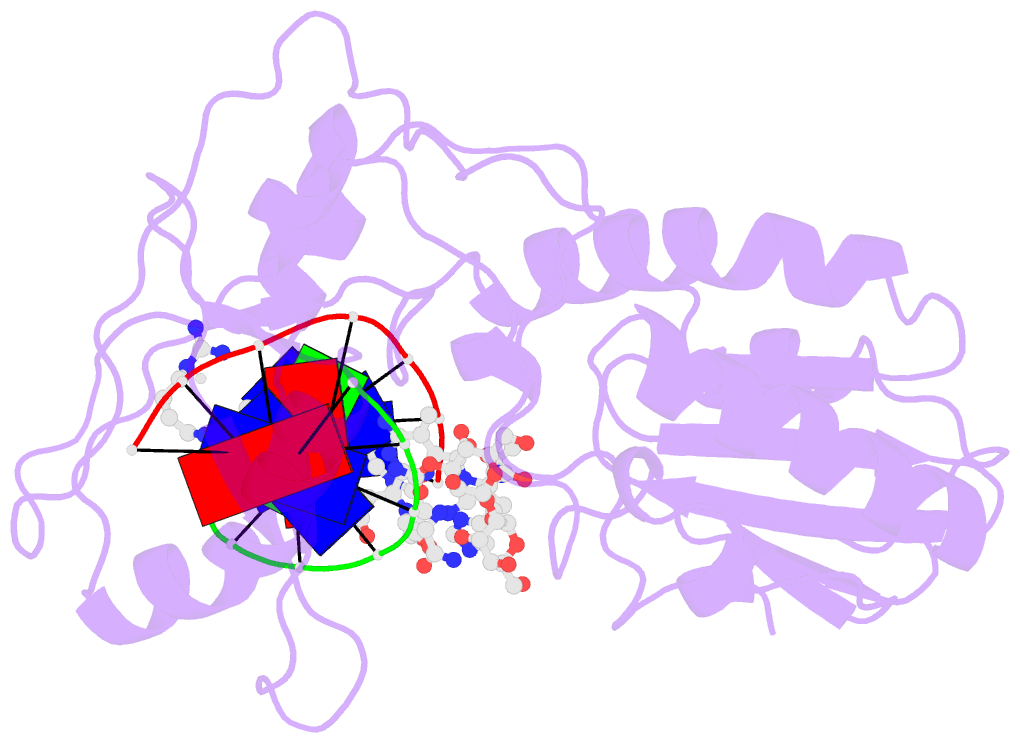
- Summary
- Co(iii)bleomycinb2 bound to d(attagttataactaat) complexed with mmlv rt catalytic fragment
- Reference
- Goodwin KD, Lewis MA, Long EC, Georgiadis MM (2008): "Crystal structure of DNA-bound Co(III) bleomycin B2: Insights on intercalation and minor groove binding." Proc.Natl.Acad.Sci.Usa, 105, 5052-5056. doi: 10.1073/pnas.0708143105.
- Abstract
- Bleomycins constitute a widely studied class of complex DNA cleaving natural products that are used to treat various cancers. Since their first isolation, the bleomycins have provided a paradigm for the development and discovery of additional DNA-cleaving chemotherapeutic agents. The bleomycins consist of a disaccharide-modified metal-binding domain connected to a bithiazole/C-terminal tail via a methylvalerate-Thr linker and induce DNA damage after oxygen activation through site-selective cleavage of duplex DNA at 5'-GT/C sites. Here, we present crystal structures of two different 5'-GT containing oligonucleotides in both the presence and absence of bound Co(III).bleomycin B(2). Several findings from our studies impact the current view of bleomycin binding to DNA. First, we report that the bithiazole intercalates in two distinct modes and can do so independently of well ordered minor groove binding of the metal binding/disaccharide domains. Second, the Co(III)-coordinating equatorial ligands in our structure include the imidazole, histidine amide, pyrimidine N1, and the secondary amine of the beta aminoalanine, whereas the primary amine acts as an axial ligand. Third, minor groove binding of Co(III).bleomycin involves direct hydrogen bonding interactions of the metal binding domain and disaccharide with the DNA. Finally, modeling of a hydroperoxide ligand coordinated to Co(III) suggests that it is ideally positioned for initiation of C4'-H abstraction.